- Clone
- 113-16 (See other available formats)
- Regulatory Status
- RUO
- Other Names
- CD27 ligand, CD27L, Ki-24 antigen, Tumor necrosis factor ligand superfamily, member 7 (TNFSF7)
- Isotype
- Mouse IgG1, κ
- Barcode Sequence
- CGCGAACATAAGAAG
- Ave. Rating
- Submit a Review
- Product Citations
- publications
| Cat # | Size | Price | Quantity Check Availability | Save | ||
|---|---|---|---|---|---|---|
| 355117 | 10 µg | £253 | ||||
CD70, also known as CD27L, is a 50 kD type II transmembrane glycoprotein and member of the tumor necrosis factor superfamily. CD70 is expressed on activated T, B and NK cells, activated plasmacytoid dendritic cells (pDCs), and chronic B cell lymphocytic leukemia and large B cell lymphomas. CD70 costimulates T cell proliferation and differentiation. It plays a role in the pDC-induced B cell differentiation. The ligand of CD70 is CD27.
Product DetailsProduct Details
- Verified Reactivity
- Human
- Antibody Type
- Monoclonal
- Host Species
- Mouse
- Immunogen
- CD70-transfected L cells
- Formulation
- Phosphate-buffered solution, pH 7.2, containing 0.09% sodium azide and EDTA
- Preparation
- The antibody was purified by chromatography and conjugated with TotalSeq™-A oligomer under optimal conditions.
- Concentration
- 0.5 mg/ml
- Storage & Handling
- The antibody solution should be stored undiluted between 2°C and 8°C. Do not freeze.
- Application
-
PG - Quality tested
- Recommended Usage
-
Each lot of this antibody is quality control tested by immunofluorescent staining with flow cytometric analysis and the oligomer sequence is confirmed by sequencing. TotalSeq™-A antibodies are compatible with 10x Genomics Single Cell Gene Expression Solutions.
To maximize performance, it is strongly recommended that the reagent be titrated for each application, and that you centrifuge the antibody dilution before adding to the cells at 14,000xg at 2 - 8°C for 10 minutes. Carefully pipette out the liquid avoiding the bottom of the tube and add to the cell suspension. For Proteogenomics analysis, the suggested starting amount of this reagent for titration is ≤ 1.0 µg per million cells in 100 µL volume. Refer to the corresponding TotalSeq™ protocol for specific staining instructions.
Buyer is solely responsible for determining whether Buyer has all intellectual property rights that are necessary for Buyer's intended uses of the BioLegend TotalSeq™ products. For example, for any technology platform Buyer uses with TotalSeq™, it is Buyer's sole responsibility to determine whether it has all necessary third party intellectual property rights to use that platform and TotalSeq™ with that platform. - Application Notes
-
Additional reported applications for the relevant formats include: blocking of plasmacytoid dendritic cell induced B cell proliferation and Ig secretion1.
- Additional Product Notes
-
TotalSeq™ reagents are designed to profile protein levels at a single cell level following an optimized protocol similar to the CITE-seq workflow. A compatible single cell device (e.g. 10x Genomics Chromium System and Reagents) and sequencer (e.g. Illumina analyzers) are required. Please contact technical support for more information, or visit biolegend.com/totalseq.
The barcode flanking sequences are CCTTGGCACCCGAGAATTCCA (PCR handle), and BAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA*A*A (capture sequence). B represents either C, G, or T, and * indicates a phosphorothioated bond, to prevent nuclease degradation.
View more applications data for this product in our Scientific Poster Library. -
Application References
(PubMed link indicates BioLegend citation) -
- Shaw J, et al. 2010. Blood 115:3051. (Block)
- Product Citations
-
- RRID
-
AB_2749965 (BioLegend Cat. No. 355117)
Antigen Details
- Structure
- Type II transmembrane glycoprotein, member of the tumor necrosis factor superfamily, 50 kD
- Distribution
-
Activated T cells and B cells, activated NK cells, activated plasmacytoid dendritic cells, some B cell lymphomas
- Function
- Role in T cell proliferation, generation of cytolytic and memory T cells, B cell proliferation and differentiation
- Ligand/Receptor
- CD27
- Cell Type
- B cells, Dendritic cells, NK cells, T cells
- Biology Area
- Costimulatory Molecules, Immunology
- Molecular Family
- CD Molecules
- Antigen References
-
1. Bowman MR, et al. 1994. J. Immunol. 152:1756.
2. Shaw J, et al. 2010. Blood 115:3051.
3. Keller AM, et al. 2009. Blood 113:5167. - Gene ID
- 970 View all products for this Gene ID
- UniProt
- View information about CD70 on UniProt.org
Related FAQs
- What are TotalSeq™ reagents and how do they work with established workflows (CITE-seq, and REAP-seq)?
-
TotalSeq™ is BioLegend’s brand of antibody-oligonucleotide conjugates, to enable simultaneous analysis of proteins and mRNA in single cells. CITE-seq and REAP-seq are two similar workflows for simultaneous protein and mRNA analysis, and the TotalSeq™ conjugates integrate seamlessly into these established protocols.
- What is the difference between TotalSeq™-A, -B, and –C?
-
The main difference between the 3 formats relates to the sequences of the oligo tags and their downstream compatibility with single-cell sequencing platforms.
- TotalSeq™-A reagents use a poly-A capture method that is instrument agnostic which includes the 10x Genomics 3’ Single Cell Gene expression kit.
- TotalSeq™-B utilizes the feature barcode technology for 10x Genomics 3’ Single Cell Gene expression kit.
- TotalSeq™-C also uses the feature barcode technology but for 10x Genomics 5’ V(D)J expression kit.
They each use different primers for library amplification. You can read a little more about the differences on our TotalSeq™ webpage. - Can I use TotalSeq™-A and -B together?
-
We are not currently able to support the mixing of TotalSeq™-A and TotalSeq™-B reagents in a single experiment. These two product lines require different protocols for library prep, and the different capturing methods may cause discrepancies in capture efficiency. In addition, it can often cause issues downstream in the bioinformatics portion when analyzing your data due to the variations in index sequences and lengths. In theory, they should work together, but in practice it can be quite difficult and we advise against doing so whenever possible.
- Can I use TotalSeq™-A on the 10x Genomics 3’ Single Cell Gene platform or do I need TotalSeq™-B?
-
Either will work fine.
- What are “features”?
-
When discussing single-cell data, features are any aspect of the cell that is being measured. Common features are mRNA, protein, sgRNA, etc.
- Is TotalSeq™-A, B, and C supported by 10x Genomics?
-
TotalSeq™-A/B/C products are fully supported by BioLegend and have been internally tested by BioLegend on the 10x Genomics platform. TotalSeq™-B and C are fully supported by 10x Genomics. 10x Genomics provides limited support for TotalSeq™-A.
If you are unsure about which technical service team to contact if you have questions or issues, feel free to reach out to BioLegend and we will work with our partners to resolve your problem or answer your technical questions. - What are "Hashtags"?
-
Hashtag reagents are intended to be used for sample barcoding, which allows users to combine multiple samples into a single lane, then de-multiplex during analysis. Instead of select antigen-specific antibodies, the hashtags are designed so that they are specific for human or mouse cells, and to cover as many cells as possible. For human samples, the hashtags are made of two antibodies that recognize ubiquitous surface markers, CD298 and β2 microglobulin, each conjugated to the same oligonucleotide containing the barcode sequence. For mouse samples, the surface markers are CD45 and H-2 MHC class I. The conjugates are already pre-mixed and ready to use.
- What’s the benefit of Cell hashing?
-
Hashing allows customers to run multiple samples in a single lane of a 10x Genomics Chromium instrument, or equivalent, which optimizes the number of cells or samples that can be analyzed simultaneously. This can reduce variability due to batch effects or sample handling. It can also help to identify doublets more efficiently. Furthermore, it can also improve yield and help with cell input when cell numbers are low, therefore optimizing the use of the platform and your workflow or protocol.
- What is “super loading”?
-
Typically, when using a single-cell platform, as you increase the amount of input cells, the rate of doublets (two cells captured as one data point) increases. This leads to an unacceptable percentage of datapoints that contain more than one cell. Hashtags allows you to “super load” the number of cells. To this end, multiple aliquots of the same sample could be stained with as many hashtags as aliquots you’d wish to mix. The number of aliquots and number of cells per aliquot may vary, but after washing and mixing the cells, following the staining and analysis protocol, if more than one hashtag is detected in a single-cell “event”, that event can be safely discarded as a doublet. This does not eliminate doublets that contain the same hashtag but the rate of doublet detection is greatly increased. However, “super loading” should be carefully controlled as the more cells you load the more doublets occur, causing diminishing returns overall. There should be a balance between multi-sample optimization, single-cell data yield, and optimal instrument/platform performance.
- What are nuclear hashtags?
-
Nuclear hashtags are used for single-nucleus RNA-seq (snRNA-seq) samples. snRNA-seq captures RNAs that are isolated with the nucleus. This is done because whole, intact cells may be difficult to isolate due to specific experimental or sample conditions, or to answer a specific scientific question. See an example of nuclear hashtag application in this paper: https://www.nature.com/articles/s41467-019-10756-2
- Can I use nuclear hashtags for single cell ATAC-seq?
-
Unfortunately, our TotalSeq™ reagents are not directly compatible with single ATAC-seq kits at this time. However, the NYGC has pioneered a bridging method ATAC with Select Antigen Profiling by sequencing (ASAP-seq) which is able to bridge TotalSeq™ antibodies with other capture platforms such as scATAC-seq. https://www.biorxiv.org/content/10.1101/2020.09.08.286914v1
Our nuclear hashtag products available off the shelf, but we do not currently have a recommended protocol for this application at this time.
The only recommendations we can provide are literature references such as this one:
https://www.nature.com/articles/s41467-019-10756-2 - Can I get technical support when using hashtags?
-
Hashtags can absolutely be used in any single cell platform including the 10x Genomics platform. BioLegend fully supports the usage of hashtags. Please contact our technical service group for more information.
- Does co-staining for FACS and TotalSeq™ interfere with the workflow?
-
We have not tested this, but there is no reason to believe that non-competing clones for the same target would be problematic. Similarly, a sub-saturating concentration of both reagents against the same target should be tolerated by the cell/technique. The original CITE-seq paper by Stoeckius et al.(Fig. 2) demonstrated that cells can be co-stained for downstream analyses. Although the authors did not use current TotalSeq™ reagents, the technology is the same. In addition, other CITE-seq users have also demonstrated this approach. Please contact our technical service group for more information.
- Why is it essential to remove dead cells prior to subjecting samples for CITE-seq?
-
We recommend >95% viability before beginning you CITE-seq experiment. Running dead cells during CITE-seq essentially leads to “wasting” single cell runs on dead cells, which may ultimately lead to sub-optimal data, or not processing enough cells to pick up the positive events, especially if the frequency of your target cell is very low. Dead cells can also be removed using magnetic beads. If performing CITE-seq before eliminating dead cells is required, it is possible to use bioinformatics methods to try to clean up low quality events (which may include dead cells). This could be a more complex approach, but in such cases, we recommend the following reading:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4758103/
https://www.ncbi.nlm.nih.gov/pubmed/28045081 - Is it necessary to sort samples prior to CITE-seq? What should I consider when deciding to pre-sort, or not?
-
If the populations of interest can be clustered/identified without sorting, the likelihood that you need to pre-sort is minimal. However, if the cell number in the cluster is very small, it may not be sufficient to draw conclusions when compared to control, or other cell populations. In which case, enrichment of this population prior to CITE-seq analysis may help with obtaining more meaningful sequencing data that is selective for your rare cell population(s) of interest. We also recommend to sort out dead cells if the viability in a sample is lower than 95%.
- How can I titrate TotalSeq™ antibodies?
-
We have developed pre-titrated, lyophilized TotalSeq™ panels to facilitate the use of combined antibodies. We have panels in all three formats (TotalSeq™-A, B, and C) designed to identify the main immune cells, as well as a universal panel that contains antibodies against 130 protein targets, and 7 isotype controls. We will keep on releasing these pre-optimized panels as we understand that titrating dozens of antibodies for this application is not an easy task. We may not be able to provide specific titration values for individual antibodies as that may depend on multiple factors. However, here are some recommendations in case you may not be able to use our panels, and need to perform your own titrations:
- Performing titrations using the actual CITE-seq method is costly, but using hashtags can make the process more affordable. See Fig. 3 and associated methods of this publication for more information.
- Perform flow cytometry experiments to find the optimal antibody concentration, using the same clone conjugated to PE. The flow cytometry antibody amount should translate well to CITE-seq.
- Label the cells with different concentrations of a TotalSeq™ antibody, and then use a poly(dT) oligo conjugated to a fluorophore, such as Alexa Fluor® 647, as a secondary reagent to detect the TotalSeq™ antibody. This is a more direct method, but it requires the use of the actual TotalSeq™ antibody.
- What tools are available for data analysis?
-
Some available resources include CITE-Seq-Count, and the tools developed by the Rahul Satija laboratory. Please contact our technical service group for further information.
- I need to order primers; where do I order them and what purity is needed?
-
There are several companies offering primer synthesis. We can recommend IDT technologies. For additive primers, desalt purification is sufficient. For index primers, either HPLC or PAGE is needed.
- Why should I generate ADT/HTO libraries separate from mRNA?
-
Separate ADT/HTO and mRNA libraries provide flexibility when sequencing. Libraries can be mixed at different ratios before sequencing depending on the number of reads needed. Typically, ADT/HTO libraries require less sequencing depth compared to their mRNA counterparts.
- How is the oligo tag bound to the antibody so that it does not interfere with antibody binding to target protein?
-
The oligo is attached to the antibody in the same method as some of our fluorophore-conjugated antibodies that are quality tested by flow cytometry. Once the antibodies have been conjugated to the oligonucleotide, we verify by flow cytometry that the antibody can still bind to its target. This is part of our quality control process for TotalSeq™ conjugates.
- Is it possible to run CyTOF® and TotalSeq™ (CITE-seq) alongside each other?
-
Not in the same experiment; it is not possible because CyTOF® requires its own instrument and protocol, and it destroys the sample in the process. To do so, it is necessary to split the sample and run both CyTOF® and CITE-seq assays. Note that CITE-seq generates equivalent data for surface proteins as CyTOF® with higher panel multiplexing capabilities and at a single-cell level.
- Can CITE-seq be extended to micro-RNAs?
-
This is a technically challenging application as micro-RNAs are very small, and in many cases will be as small as or even smaller than the required primers to execute the protocol. It is not possible at the moment.
- Can I use TotalSeq™ in bulk RNA-seq experiments?
-
Yes, the protocol to perform bulk epitope and nucleic acid sequencing (BEN-seq) has been developed as a collaboration between Illumina and BioLegend. You can read more about it in our application note.
- Is autofluorescence an issue as in flow cytometry?
-
No, sequencing as the final readout is not affected by cell autofluorescence.
- What is the baseline copy number for detection of protein expression (via TotalSeq™ labeling)?
-
This has to do mostly with antibody affinity, titration, and level of antigen expression. BioLegend and some collaborators are working on titration data. There is also some data already published. In theory, as long as the antibody can bind one molecule, PCR amplification and sequencing should pick it up, but there is always background noise. A common way to control for this is to add negative cells to your experiment. For example, if working with human PBMCs, use mouse cells and vice versa.
- How many cells do you need per experiment?
-
This depends on the biological question that needs to be answered. From the technical perspective, when using 10x Genomics Chromium, it is recommended to load 5,000 – 10,000 cells per lane. The use of hashtags can increase that number. For ease of staining, we typically recommend 1 million cells, but this number also depends on availability of cells and the operator’s ability to handle low cell numbers.
- How many TotalSeq™ antibodies is it possible to multiplex?
-
We have optimized multiple pre-mixed antibody cocktails, including TotalSeq™-A, -B, or -C Human TBNK that contain 9 antibodies and our TotalSeq™-C Human Universal Cocktail, v1.0 which contains an unprecedented 130 target antibodies plus 7 isotype controls in a single vial. We will be releasing more panels in the near future. The literature suggests that panels larger than these cocktails are possible. For example, Yapeng et al. published a study using 197 TotalSeq™ antibodies and 7 isotype controls. BioLegend and the scientific community keep pushing the upper limits of multiplexed protein detection by sequencing and have yet to find one.
- Is it possible to do FACS sorting first then do CITE-Seq, while performing both antibody-oligo and antibody-fluorophore labelling at the same time? Will the oligo conjugate withstand sorting?
-
The oligo should withstand sorting. However, based on theoretical considerations we recommend using non-competing clones, to minimize impact in either FACS or sequencing signal/reading. Even if exposed for a very short time, we have not evaluated the effect of ultraviolet or violet lasers on the oligo conjugates. An alternative to flow sorting could be negative magnetic bead enrichment, such as our MojoSort™ platform.
- What should be my sequencing depth be for my ADT/HTO library?
-
We recommend a sequencing depth of 5,000 reads/cell. If you are using more than 100 TotalSeq™ antibodies, you should increase to 10,000 reads/cell. For HTOs alone, 500 reads/cell is sufficient.
- Do I need isotype controls?
-
Isotype controls are highly recommended but not required. The utility of isotype controls in these applications is that they can help identify “sticky cells” caused by cells with compromised viability. Isotype controls also provide an overall picture of the background level or non-specific binding of different cell types. Typically when running isotype controls for CITE-seq assays, you want to include 1 isotype control for every isotype included in the panel, regardless of how many antibodies there are. We recommend using the isotype control at a concentration matching the highest concentration of a single antibody used in a panel, for that particular isotype. Typically, the isotype controls should be used in the same tube as the panel itself. Unlike a traditional flow cyometry experiment, the isotype controls for multiomics cell characterization do not need their own separate samples for comparison. Isotype controls are included in our TotalSeq™ Universal Cocktails.
Another useful control to help identify positive signals are cells known to not bind the antibodies used. For example, murine cells when characterizing human samples, as long as the antibodies do not cross-react between mouse and human targets. This, however, is not required to confidently identify positive signals. - Should I use dextran sulfate to block?
-
Several genomics protocols, including some from 10x Genomics, may recommend including dextran sulfate as a blocker to prevent unwanted charge interactions between nucleic acids and other positively charged molecules. However, we have found that in some cases, the inclusion of dextran sulfate can interfere with antibody binding and recognition and we do not recommend using it with any of our TotalSeq™ reagents. It’s unclear what is the exact mechanism of action that causes this issue. All our protocols reflect this observation; thus we do not recommend the use of dextran sulfate.
Other Formats
View All CD70 Reagents Request Custom Conjugation| Description | Clone | Applications |
|---|---|---|
| Purified anti-human CD70 | 113-16 | FC |
| PE anti-human CD70 | 113-16 | FC |
| FITC anti-human CD70 | 113-16 | FC |
| PerCP/Cyanine5.5 anti-human CD70 | 113-16 | FC |
| APC anti-human CD70 | 113-16 | FC |
| Alexa Fluor® 647 anti-human CD70 | 113-16 | FC |
| PE/Cyanine7 anti-human CD70 | 113-16 | FC |
| Biotin anti-human CD70 | 113-16 | FC |
| TotalSeq™-A0027 anti-human CD70 | 113-16 | PG |
| TotalSeq™-C0027 anti-human CD70 | 113-16 | PG |
| APC/Fire™ 750 anti-human CD70 | 113-16 | FC |
| PE/Dazzle™ 594 anti-human CD70 | 113-16 | FC |
| TotalSeq™-B0027 anti-human CD70 | 113-16 | PG |
| TotalSeq™-D0027 anti-human CD70 | 113-16 | PG |
Compare Data Across All Formats
This data display is provided for general comparisons between formats.
Your actual data may vary due to variations in samples, target cells, instruments and their settings, staining conditions, and other factors.
If you need assistance with selecting the best format contact our expert technical support team.
-
Purified anti-human CD70
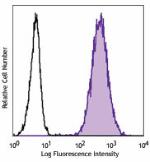
Human myeloma cell line, U266, was stained with purified CD7... -
PE anti-human CD70
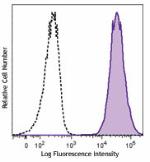
Human myeloma cell line, U266, was stained with CD70 (clone ... -
FITC anti-human CD70
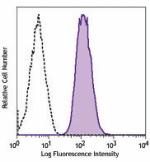
Human myeloma cell line, U266, was stained with CD70 (clone ... -
PerCP/Cyanine5.5 anti-human CD70
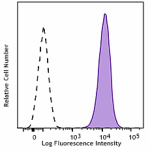
Human myeloma cell line U266 was stained with CD70 (clone 11... -
APC anti-human CD70
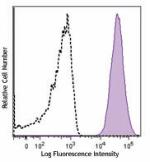
Human myeloma cell line, U266, was stained with CD70 (clone ... -
Alexa Fluor® 647 anti-human CD70
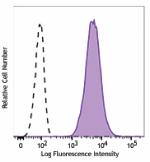
Human myeloma cell line, U266, was stained with CD70 (Clone ... -
PE/Cyanine7 anti-human CD70
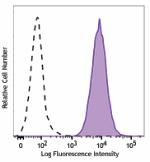
Human myeloma cell line, U266, was stained with CD70 (clone ... -
Biotin anti-human CD70
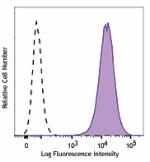
Human myeloma cell line, U266, was stained with biotinylated... -
TotalSeq™-A0027 anti-human CD70
-
TotalSeq™-C0027 anti-human CD70
-
APC/Fire™ 750 anti-human CD70
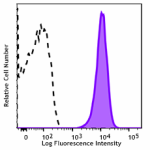
Human myeloma cell line, U266, was stained with CD70 (clone ... -
PE/Dazzle™ 594 anti-human CD70
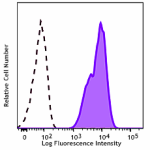
Human myeloma cell line, U266, was stained with CD70 (clone ... -
TotalSeq™-B0027 anti-human CD70
-
TotalSeq™-D0027 anti-human CD70
 Login / Register
Login / Register 













Follow Us