Autophagy is a process that degrades and removes dysfunctional proteins, damaged organelles, and intracellular pathogens by delivering cytoplasmic material to the lysosome. A primary and unique function of autophagy is to degrade entire organelles, which can be used to recycle cell components and generate energy. In conditions of starvation, autophagy is boosted to produce amino acids and ATP for cell survival, but in the presence of growth factors, autophagy pathways are shut off. Dysregulation of autophagy can have fatal consequences for cells, which is why it has been linked to diseases like cancer, neurodegeneration, and pathogenic infections.
Autophagy
Download our autophagy poster to learn more
Autophagy Pathways and Regulation
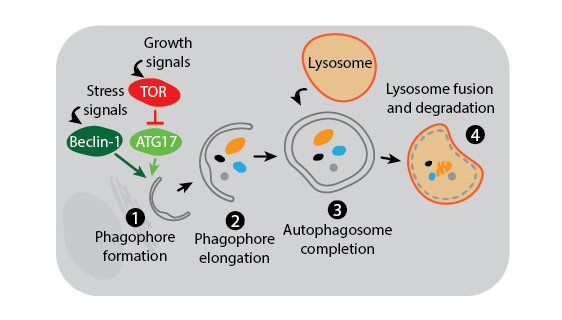
Three main autophagy pathways have been described: macroautophagy, microautophagy, and chaperone-mediated autophagy. The process and regulation of macroautophagy, which is the primary and most studied autophagy pathway, is depicted in the figure below.
Macroautophagy. Macroautophagy activation is under the control of multiple signaling mechanisms, two of which are shown in the above figure. In the presence of growth signals, TOR kinase suppresses autophagy initiation by inhibiting a kinase complex containing ATG17. In contrast, stress signals induce activation of Beclin-1 to trigger formation of the phagophore membrane which is likely derived from the endoplasmic reticulum (1). Many other autophagy proteins are then involved in the downstream process of creating and elongating the phagophore (2), joining the phagophore ends to complete the phagosome around cytoplasmic contents (3), and finally fusing the phagosome with the lysosome for degradation (4).
Microautophagy. Microautophagy refers to the direct engulfment of cytoplasmic content by the lysosome. There is evidence for several distinct types of microautophagy that differ based on the formation of the membrane, which can invaginate or protrude around cellular components. Similar to macroautophagy, microautophagy can be selective or non-selective, is able to degrade entire organelles, and can be activated by signals like environmental stress.
Chaperone-mediated Autophagy. Chaperone-mediated autophagy utilizes heat shock proteins and transporters on lysosomal membranes to degrade proteins that express a targeting motif, making it a highly specific process. The targeting motif is recognized by chaperones like HSPA8/HSC70, which transfer the protein to the lysosomal membrane. Upon binding with LAMP-2A on the lysosome, the target is internalized into the lysosomal lumen for degradation. Chaperone-mediated autophagy is regulated by environmental stress, and LAMP-2A expression levels can dictate the rate at which it occurs.
Tools for Studying Autophagy Pathways and Regulation
| Antibodies for Autophagosome Markers | Antibodies for Transcription factors Regulating Autophagy Genes | Antibodies for Autophagy Protein Trafficking |
Autophagy and Neurodegenerative Disease
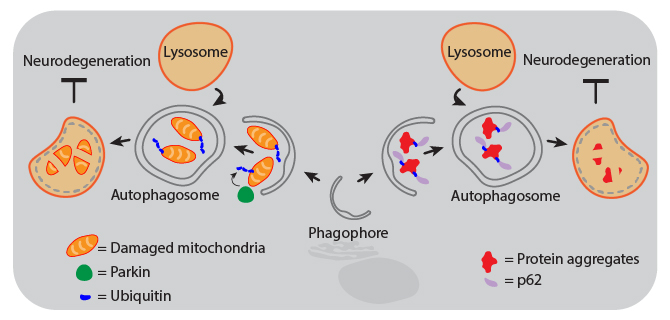
Animal and human studies have shown that autophagy is protective against neurodegeneration, and defects in autophagy pathways are associated with diseased brains. For example, the lack of ATG5 or ATG7 results in neurodegeneration of the CNS in mice. Decreased autophagic function can result from gene mutations that inhibit autophagosome maturation and lysosome fusion, or from aging-related declines in CNS expression of autophagy proteins. Therapies that activate the autophagy pathways are therefore being investigated as treatments for neurodegenerative diseases. Proper activation of autophagy can maintain neuronal health through several mechanisms, including aggrephagy and mitophagy, both of which are forms of selective macroautophagy.
| Mitophagy. Mitochondrial dysfunction can drive pathogenesis of neurodegenerative diseases like Parkinson’s. Removal of damaged mitochondria by autophagy is called mitophagy, which is triggered by accumulation of activated PINK1 at the mitochondrial membrane. This leads to phosphorylation of Parkin, which then ubiquitinates mitochondrial components to signal the organelle for degradation. | Aggrephagy. Aggregates of proteins like α-Synuclein, Amyloid-β, and Tau can cause neurodegeneration in Parkinson’s and Alzheimer’s diseases. These protein aggregates become too large to pass through the pore of the proteasome, so cells rely on autophagy for their removal to maintain health. Following ubiquitination, protein aggregates are recognized by the adaptor protein p62 (also known as sequestosome-1, SQSTM1) that can then link the aggregates to phagophore-associated LC3, resulting in their engulfment and degradation. |
Tools for Studying the Role of Autophagy in Neurodegeneration
| Antibodies for Autophagy and Neurodegeneration | Autophagy Antibody Sampler Kit | MitoSpy™ Mitochondrial Probes |
Autophagy and Cancer
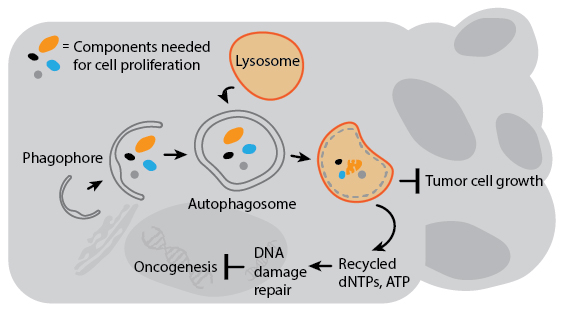
Several genes involved in tumor suppression and/or are associated with cancer susceptibility are also regulators of autophagy. Decreased expression of Beclin-1 is associated with prostate and breast cancers, and gene transfer of Beclin-1 to cancer cell lines can inhibit their growth in culture. Autophagy’s anti-cancer properties are likely attributable to mechanisms that indirectly inhibit cancer growth and spread. For example, deleting autophagy genes promotes genomic instability which can increase the chances for oncogenesis. Autophagy may also degrade organelles and cell components essential for cell growth, or inhibit the growth of tumors by preventing necrosis to avoid cancer-promoting inflammation. In the context of current cancer drug treatment, however, autophagy actually suppresses cell death in animal models. This is likely a result of autophagy’s pro-survival response to drug-induced stress. A detailed understanding of how autophagy impacts cancer will therefore be critical for the development of combination therapies that involve autophagy regulation.
Tools for Studying the Role of Autophagy in Cancer
| Immuno-oncology Research Tools | Phospho-specific Antibodies | GoInVivo™ Antibodies |
Autophagy and Immune Activation
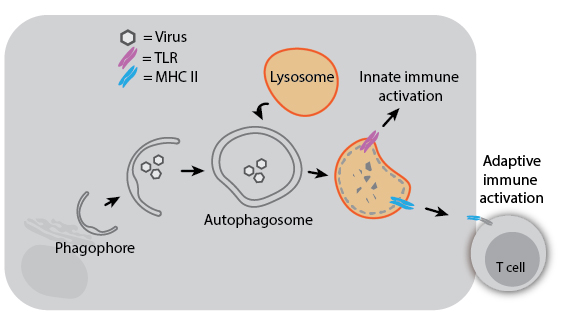
Because autophagy can degrade intracellular pathogens (referred to as “xenophagy”), it plays a role in both innate and adaptive immunity. Degradation of viruses by autophagy can aid immune responses by delivering viral components to TLRs for sensing or to MHC II proteins for antigen presentation. Herpes Simplex Virus 1 (HSV-1) even encodes a virulence factor that binds Beclin-1 to inhibit autophagy, and mutation of this viral factor severely compromises its ability to cause disease in mice. In the absence of infection, autophagy may still impact immune development and activation: T cells from ATG5-knockout mice have decreased proliferative ability, and autophagy regulatory genes have been associated with susceptibility to Crohn’s disease. Understanding the diverse roles of autophagy and its mechanisms may help us treat infectious and inflammatory diseases more effectively.
Tools for Studying the Role of Autophagy in Infection and Inflammation
| Anti-Virus LEGENDplex™ Panels | Inflammation LEGENDplex™ Panels | Flex-T™ Tetramers |
References
Levine et al. 2008 Cell. PMID: 18191218
Mizushima et al. 2007 Genes Dev. PMID: 18006683
Glick et al. 2010 J Pathol. PMID: 20225336
Deretic et al. 2013 Nat Rev Immunol. PMID: 24064518
Todde et al. 2009 Biochim Biophys Acta. PMID: 19022377
Oku et al. 2018 Bioessays. PMID: 29708272
Parzych et al. 2014 Antioxid Redox Signal. PMID: 23725295
 Login / Register
Login / Register 






Follow Us