Tips and Tricks for Titrating TotalSeq™ Antibodies
Performing titrations to determine the optimal amount of antibody is important for any antibody-based application. If you use too low of a concentration, a positive signal may not be distinguishable from a negative signal. Conversely, if you use too high of a concentration, it will increase background with the additional downside of wasting the reagent.
When using TotalSeq™ antibodies for applications like CITE-seq, it is highly recommended that you titrate to get optimal results. There are different considerations when optimizing the antibody concentration. Like other applications, using too little antibody can result in a poor separation between positive and negative populations. However, as you increase the antibody concentration, the background doesn’t typically increase. Instead, excess antibody will result in unnecessary sequencing depth, using up too many of your reads and affecting the ability to identify rare or lowly expressed targets. In this blog, we’ll provide guidance on how to titrate antibodies specifically for sequencing experiments.
TotalSeq™ Titration Methods
Ideally when titrating antibodies for CITE-seq, single-cell sequencing should be used as the readout to ensure that the titration values are appropriate for this application. However, this can be costly in terms of both time and resources. An easier and more cost-effective approach is to perform titrations using the same clone in flow cytometry. Flow cytometry titrations can provide a concentration that is similar to what is appropriate for CITE-seq. Usually these values are higher than what is required for sequencing because sequencing is more sensitive. In a sequencing experiment, the signal is amplified by PCR, and unique molecular identifiers (UMIs) control asymmetric amplification.
When titrating antibodies with flow cytometry, we recommend using PE-conjugated antibodies because they are compatible with most flow cytometry instruments and provide a clear, strong signal. It is possible to use other fluorophores. When choosing a fluor, keep in mind that a dimmer fluorophore might give a false negative signal since more antibody may be needed to be analyzed by flow cytometry compared to sequencing. If you are using flow cytometry to titrate, we advise not titrating more than one antibody at a time. Compensation and instrument configurations can make this difficult and give false positive or negative signals.
Titration of our hashtag antibodies has different considerations because each reagent is provided as a mix of two antibody clones. To titrate your hashtags, we recommend using a PE-conjugated format of both clones (for mouse, clones 30-F11 and M1/42, and human, clones LNH-94 and 2M2). Combine these clones at a 1:1 concentration for testing. Note, our PE-conjugated anti-human antibodies are provided at a test size so this may not be the same volume for each antibody. Please refer to the certificate of analysis to determine the concentration of each lot.
Another flow cytometry-based titration technique is to use fluorescently-tagged oligonucleotides that will hybridize to the TotalSeq™ antibody oligonucleotide tag. The sequence of the oligonucleotide that is fluorescently conjugated should be complementary to the TotalSeq™ oligo tag and will vary depending on which format of TotalSeq™ antibody is used. While this approach allows you to titrate the exact lot of the TotalSeq™ clone, it is less economical when compared to using flow cytometry antibodies.
Titration Workflow
The process of performing a titration of an antibody is a relatively simple process – it’s essentially just performing multiple antibody stainings in decreasing concentrations. Titrations should always be performed using the same buffer and blocking conditions that will be used when performing CITE-seq. We typically recommend beginning with ≤ 1 µg per 1 million cells. In practice, 1 µg is often the maximum amount needed to stain cells for CITE-seq; usually, it’s much lower. So, if you’re starting with 1 µg/million cells, we suggest only performing further dilutions. Known optimal amounts of antibodies for a particular clone, marker, and/or cell type for flow cytometry are also a good place to begin. The optimal concentration may vary from a known flow cytometry value because a clone’s detection may be different in a multicolor flow panel. This is due, in part, to instrument adjustments made to enable compatibility with other colors/clones and blocking/buffer conditions. If you are starting with a known concentration from previous data, use this as a midpoint as you test higher and lower concentrations around it.
Our in-house titrations normally test around 5-6 concentrations (Fig. 1), using 1:1 serial dilutions (e.g. if your starting amount of antibody is 1 μg, you’d also test 0.5 µg, 0.25 μg, 0.125 µg, 0.0625 μg, etc.). When titrating TotalSeq™ hashtags, they usually need to be titrated to lower concentrations given their high sequencing signal. This high signal occurs because hashtags antibodies are designed to recognize ubiquitous markers which are often abundantly expressed.
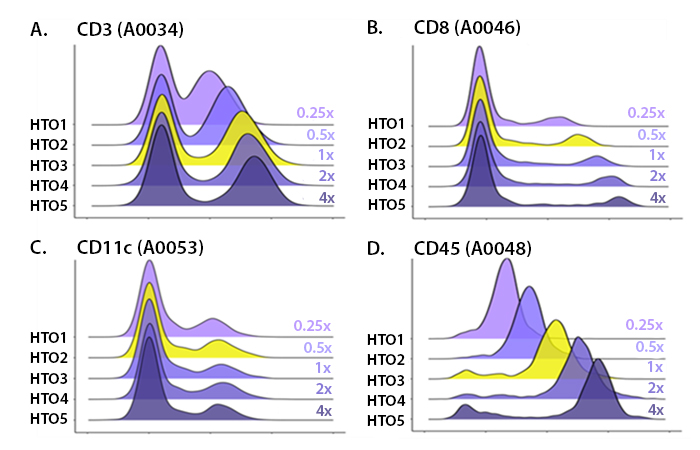
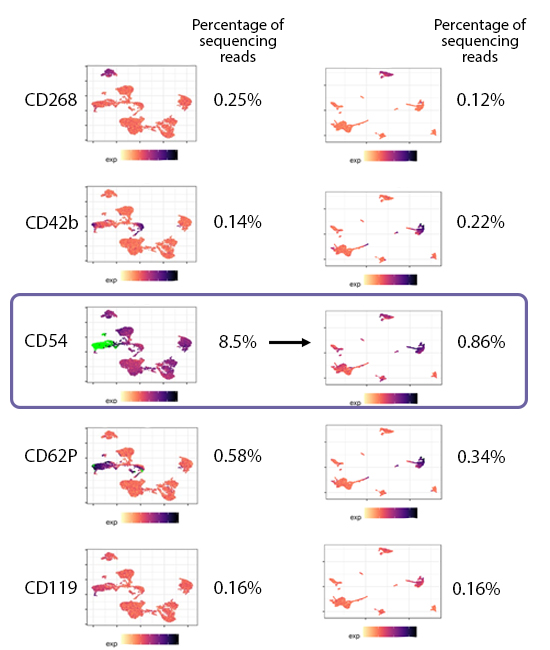
When choosing a concentration, you should utilize the minimal amount of antibody that still achieves separation of the negative and positive populations. Adding more antibody than needed will typically still separate the positive and negative populations (Fig. 1), but use up more sequencing reads. By choosing the lowest concentration required for clear separation of the positive and negative peaks, you also get the minimum needed reads to sequence. This helps reduce the possibility that you waste sequencing reads on a particular marker (Fig. 2). Instead, these reads can be utilized to identify more rare markers. If a particular marker uses a large portion of the reads, there may not be enough sequencing space to identify markers that are more difficult to detect, such as lowly abundant markers or those found on rare cell populations.
Hashtag antibodies also need to be titrated to balance the reads per each hashtagged sample. If a particular sample has more hashtag reads than another sample when multiplexed, the sample with the lower signal may begin to look like background and become harder to demultiplex. This becomes paramount as you increase the number of hashtags used because the fraction of reads per hashed sample is decreasing.
Antibody Cocktails
These titration techniques can be employed when using single antibodies (or hashtags), building a larger panel, or adding more (drop-in) antibodies to BioLegend’s pre-titrated cocktails. Removing the need for optimization, we’ve released a number of pre-titrated antibody cocktails. We provide a human TBNK cocktail in TotalSeq™-A, -B, and -C formats as well as our TotalSeq™-A Human Universal Cocktail, v1.0 and TotalSeq™-C Human Universal Cocktail, v1.0. We’ve also partnered with Mission Bio to develop the TotalSeq™-D Human Heme Oncology Cocktail, v1.0 for the co-detection of SNVs/indels, CNVs and proteins on the Mission Bio Tapestri Platform. All of these cocktails have been titrated using next-generation sequencing as a readout. Using these cocktails also minimizes batch effects from making your own panels. As an added bonus in larger panels, there are so many antibodies present that they almost act as a “blocking agent” and help minimize non-specific binding.
Titrating antibodies for CITE-seq or other sequencing applications is critical for achieving optimal results. While titrating oligo-conjugated antibodies can seem complicated, many of the same principles used for other antibody-based applications can be applied. Understanding how to select the right concentration and how to use alternatives, like PE-conjugated antibodies, to titrate can help make the process simpler. With the right optimization, TotalSeq™ reagents transform single-cell sequencing experiments with unparalleled protein detection. If you have additional questions on titration, please contact our Technical Services group.






Follow Us