There are an ever-increasing number of multiplexed antibody-based imaging methods being developed as commercial products and as community-based tools. IBEX was created to enable simple implementation of highly multiplexed spatial analysis by biologists in standard laboratory environments. IBEX is a completely open community-developed technique that is not limited to company-specified antibodies, nor does it require proprietary reagents, specialized equipment, or vendor-specific commercial software for analysis.
IBEX Protocol
The protocol and various implementations of both manual and automated IBEX on different imaging systems is described in detail in Nature Protocols. Additional details on how to apply IBEX with tissues from fluorescent reporter animals and heavily fixed tissues can be found in PNAS (Figure S4). Here we will discuss some initial reagent considerations for transitioning to IBEX from traditional single-cycle multiplexed immunofluorescence microscopy. Briefly, IBEX can be implemented at almost any scale and is compatible potentially with any tissue or sample processing method with appropriate validation and optimization.
Generally, experience with legacy techniques like chromogenic or single-cycle multiplexed fluorescent immunohistochemistry will be useful for IBEX adoption. Antibody validation and titration are crucial for establishing confidence and rigor in a new IBEX experiment. A Nature Methods primer by HuBMAP provides additional resources and guidelines for multiplexed antibody-based imaging. The technique and antibody panels as currently described use fixation with a low percentage of paraformaldeyhde followed by cryopreservation. Implementations of the method using fresh frozen or FFPE are also possible with further validation and optimization.
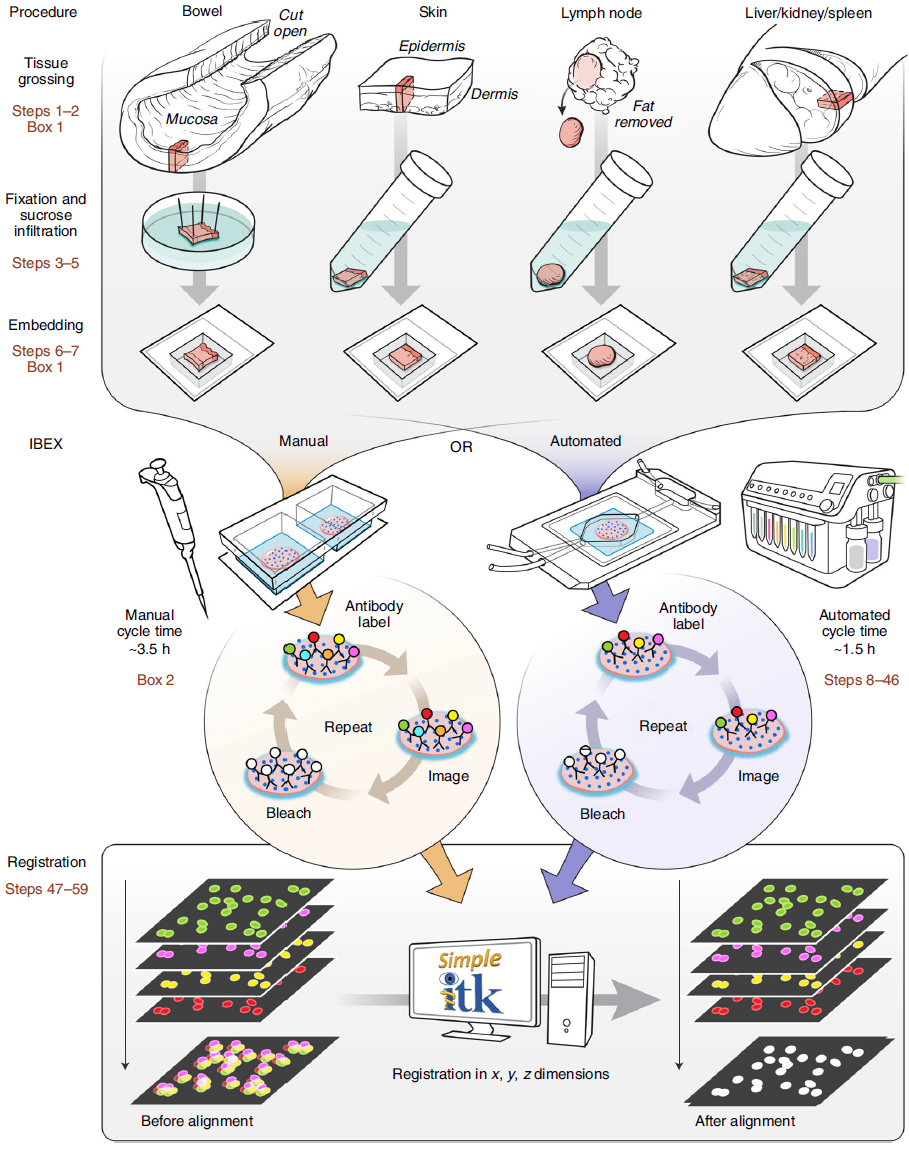
Image adapted from Nature Protocols; Figure designed by Li Yao.
Fluorophores in IBEX
The ability to image the same fluorescent channels in iterative cycles is achieved by bleaching fluorescent signal from the previous antibody panel using Lithium Borohydride (LiBH4), a strong reducing agent. Please carefully review all safety instructions before implementation. LiBH4 is capable of rapidly eliminating the fluorescent emission of many familiar dyes such as Alexa Fluor® 488, Alexa Fluor® 532, Phycoerythrin (PE), Alexa Fluor® 555, Alexa Fluor® 594, Alexa Fluor® 647, and Alexa Fluor® 700. Certain fluorophores (JOJO-1, FITC, Hoechst, Alexa Fluor® 594) require a longer incubation time or bleaching in the presence of light (BV421™ and BV510™) for signal removal. This allows different markers to be strategically placed on either LiBH4 labile or resilient dyes to remain in one or more cycles as desired by the user.
| Antibody Format | Time to Bleach (Minutes) | Relative Brightness (Brightest = 5) |
Special Considerations | |
|---|---|---|---|---|
| LiBH4 | LiBH4 + Light | |||
| BV421™* | - | <15 | 4 | Light inactivation is challenging for large sections Competes with Hoechst 33342 if being used for nuclear fiducial Endogenous autofluorescence can pose challenges, especially in human tissues |
| BV510™* | - | <15 | 1 | Light inactivation is challenging for large sections Competes with Hoechst 33342 if being used for nuclear fiducial Endogenous autofluorescence can pose challenges, especially in human tissues |
| Alexa Fluor® 488 | <15 | - | 4 | Pair with moderate to highly expressed markers, relatively versatile |
| FITC | 30 | - | 3 | Alexa Fluor® 488 preferred over FITC for IBEX |
| Alexa Fluor® 532 | <15 | - | 3 | Challenging due to spectral overlap with Alexa Fluor® 555 and Alexa Fluor® 488 |
| PE | <15 | - | 5 | Prone to photobleaching and vignetting (See Figure S1 Nature Methods) |
| Alexa Fluor® 555 | <15 | - | 3 | Pair with moderate to highly expressed markers, relatively versatile |
| Alexa Fluor® 594 | >120 | - | 5 | Can be paired with a structural marker and used as fiducial or placed in the final IBEX cycle |
| iFluor™ 594 | <15 | - | 4 | Bleachable alternative to Alexa Fluor® 594 |
| Alexa Fluor® 647 | <15 | - | 5 | Pair with dim to moderately expressed markers |
| Alexa Fluor® 700 | <15 | - | 1 | Pair with highly expressed markers and place in early imaging cycles (See Figure S1 Nature Methods) |
* The use of Brilliant Violet™ dyes in IBEX requires the addition of a light source during the bleaching step. Many other formats are compatible with IBEX, and, potentially more that are untested.Please see Table S3 Nature Protocols for additional details. Note that fluorophore brightness will vary based on application and equipment used. For comparison, you can review BioLegend’s Fluorophore Brightness Index and this helpful webpage on fluorophore brightness in microscopy.
Compatibility of a wide range of fluorophores with IBEX makes the technique flexible enough to adapt to a variety of imaging platforms, different sample types, and experimental designs. However, the selection of format for each antibody/antigen, and their relative placement among several cycles of staining/imaging/bleaching will be unique to each project. This is one of several variables that will need optimization when designing a new IBEX experiment. Additional details on the development and validation of multiplexed imaging panels can be found in Nature Methods, particularly Figure 3 and in a detailed Nature Protocols employing the multiplexed imaging platform t-CyCIF.
Choice of fiducial markers for registration of images between different cycles also influences how the antigens will be imaged throughout the experiment. Alexa Fluor® 594 may be appropriately paired with a cellular marker in some tissue types as a fiducial since it is resilient to several cycles of bleaching. However, using an antibody conjugate as a fiducial is contingent on a suitable antigen and antibody being available in the tissue/cells of interest to allow alignment of multiple images. The nucleic acid stain Hoechst 33342 can serve as an imaging fiducial and has the added benefit of assisting in nuclear segmentation. For more details on use of a fiducial for image alignment as well as open-source software options for analysis, please see this PNAS publication.
Fluorophore selection may also be determined by the availability of directly conjugated antibodies that have utility for the experiment. Some formats have a more restrictive set of commercially available clones. In addition, relative fluorophore brightness will not always predict the performance of a specific conjugated antibody. If purified formats are required due to reagent availability, secondary anti-immunoglobulin staining will need to be used in earlier cycles of the experiment (Cycles 1-2) and then the free sites blocked. This is because antibodies are not physically removed from the tissue with IBEX.
The open source image registration software developed to align IBEX-generated images can be utilized without programming experience. The software developed by Dr. Ziv Yaniv and Bradley Lowekamp of the Bioinformatics and Computational Bioscience Branch of NIAID, NIH is freely available from GitHub, along with usage instructions (for installation instructions also read the README.md file which is part of the zip file available from GitHub). Additional details can be found in the XT Register Same Channel Simple ITK Imaris Python Extension YouTube tutorial. Sample data can be accessed on Zenodo for usage of the software.
The original method and extensive Nature Protocols publication describing IBEX in detail, as well as related content can be reviewed below:
- Radtke, Andrea J et al. “IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues.” Proceedings of the National Academy of Sciences of the United States of America vol. 117,52 (2020): 33455-33465. doi:10.1073/pnas.2018488117
- Radtke, Andrea J et al. “IBEX: an iterative immunolabeling and chemical bleaching method for high-content imaging of diverse tissues.” Nature Protocols vol. 17,2 (2022): 378-401. doi:10.1038/s41596-021-00644-9
- Hickey, John W et al. “Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging.” Nature Methods vol. 19,3 (2022): 284-295. doi:10.1038/s41592-021-01316-y
- Radtke Andrea J., et al. (2021). Accompanying dataset for: "IBEX: An iterative immunolabeling and chemical bleaching method for high-content imaging of diverse tissues" [Data set]. Zenodo. doi.org/10.5281/zenodo.5244551
- Du, Z., Lin, JR., Rashid, R. et al. Qualifying antibodies for image-based immune profiling and multiplexed tissue imaging. Nat Protocols 14, 2900–2930 (2019). doi.org/10.1038/s41596-019-0206-y
- Quardokus, EM, et al. Organ Mapping Antibody Panels: a community resource for standardized multiplexed tissue imaging. Nat Methods 20, 1174-1178 (2023). doi.org/10.1038/s41592-023-01846-7
- Gola, A., Dorrington, M.G., Speranza, E. et al. Commensal-driven immune zonation of the liver promotes host defense. Nature 589, 131–136 (2021). doi.org/10.1038/s41586-020-2977-2
- Speranza, E. et al. “Age-related differences in immune dynamics during SARS-CoV-2 infection in rhesus macaques” Life Science Alliance Jan 2022, 5 (4) e202101314: doi.org/10.26508/lsa.202101314
- Madissoon, E. et al. “A spatial multi-omics atlas of the human lung reveals a novel immune cell survival niche”. bioRxiv 2021.11.26.470108; doi.org/10.1101/2021.11.26.470108
- Radtke, A. et al. “A multi-scale, multiomic atlas of human normal and follicular lymphoma lymph nodes”. bioRxiv 2022.06.03.494716; doi.org/10.1101/2022.06.03.494716
 Login/Register
Login/Register 






Follow Us