Deciphering the Immune Response to COVID-19 With Single-Cell Multiomics
Single-cell multiomics provides a complete profile of immune cell diversity. By utilizing 5’ sequencing, you can pair full-length T and B cell receptor sequencing with analysis of the transcriptome and cell surface protein expression. In this approach, single cells are separated, sequenced, indexed, and bioinformatics methods map these sequences to link BCR and TCR sequences to antigen specificity. Understanding both T and B cell repertoire diversity and the heterogeneity of cells within a sample provides important information about the responses to viruses and other pathogens, like SARS-CoV-2.
Our scientists developed TotalSeq™-C antibodies specifically for this approach. Each antibody is compatible with 10x Genomics Chromium Single-Cell Immune Profiling Solution (5’) for immune repertoire profiling of T and B cell receptors and seamlessly integrates into the workflow. Learn more about the differences between our TotalSeq™ antibody formats.
In this blog, we’ll highlight ways that COVID-19 researchers are using our TotalSeq™-C antibody in conjunction with TCR/BCR repertoire profiling to build complete immune cell profiles and uncover the detailed B cell response following infection.
Watch an on-demand webinar to learn how Dr. Yapeng Su used single-cell multiomics to study COVID-19.
The Coordinated Immune Response to COVID-19
An understanding of the coordinated immune response to COVID-19 contributes to the continued efforts to develop therapies and measure vaccine efficacy. In this Nature Medicine article, Stephenson et. al. used multiomics techniques to simultaneously analyze the single-cell transcriptome, surface proteome, and the T and B antigen receptor profile from patients with varying severities of COVID-19. They analyzed samples from 130 patients and isolated over 780,000 peripheral blood mononuclear cells to fully understand the immune response, including rare cell populations.
In patients with COVID-19, they observed an expansion of megakaryocyte progenitors, an increase in CD34+ progenitor cells primed for megakaryopoiesis, and elevated platelet activation. They also observed a population of nonclassical monocytes which may function to replenish alveolar macrophages.
In the lymphocyte compartment, there was a correlation between severe disease and clonal expansion of CD8+ T cells with an increased ratio of effector T cells to memory T cells. In asymptomatic or donors with mild disease, there was an enrichment of follicular helper and type 1 helper T cells. Interestingly, the data also showed a decrease in IgA2 and diminished IFN-α response in patients with severe disease as compared to those with mild disease. Lastly, immune repertoire profiling revealed differential BCR clonality and mutation frequencies among gender groups. Using these datasets, further studies may determine how these changes lead to different clinical outcomes.
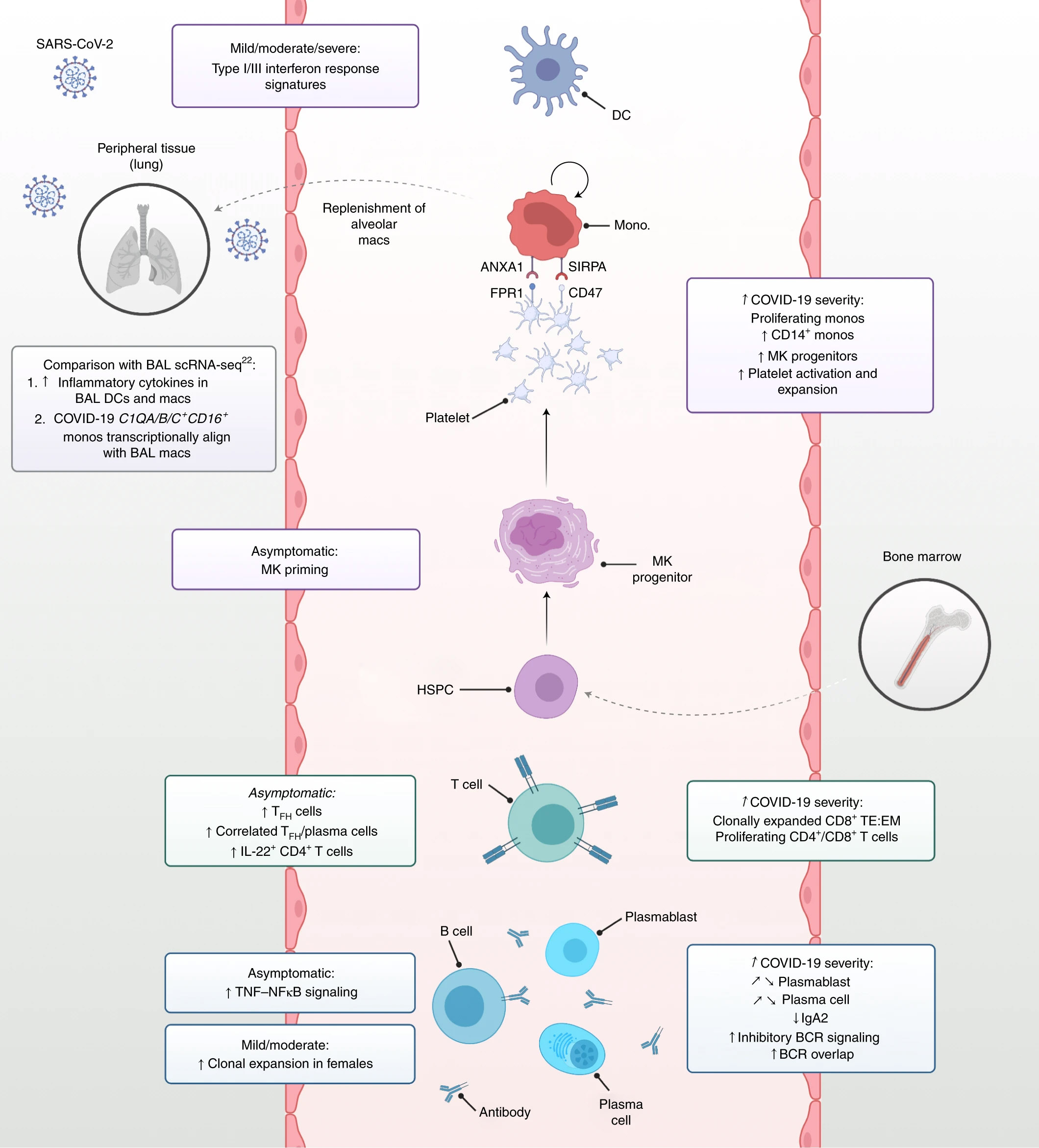
Summary of immune response profiling to COVID-19. Stephenson et al. Nat Med 27, 904–916 (2021). (CC BY 4.0)
Revealing the Anti-SARS-CoV-2 Memory B Cell Response
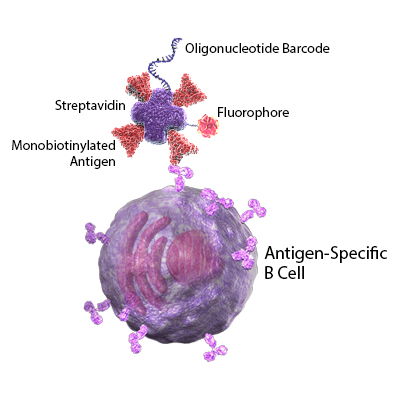
In addition to vaccine development, many researchers are working to better understand the mechanisms and duration of protective immunity in recovering patients. In this Cell publication, Sokal et al. performed longitudinal studies to profile memory B cells in patients with mild and severe cases of COVID-19. All samples were first analyzed for the presence of SARS-CoV-2 specific antibodies by ELISA. Next, they used tagged recombinant SARS-CoV-2 spike proteins to single-cell sort antigen-specific B cells. The B cells were stained for known B cell markers using TotalSeq™-oligo conjugated antibodies. The antibody-derived tags, RNA, and VDJ profiles were sequenced in single cells.
Immediately following SARS-CoV-2 infection, the authors found that existing memory B cells against other seasonal coronaviruses contribute to the immune response; however, this cross-reactive memory B cell response is short-lived. Cross-reactive clones decreased 3 to 6 months after infection and were not positively selected in the memory B cell pool. Instead, SARS-CoV-2 spike protein-specific memory B cells expanded, particularly in patients with severe disease. Levels of these spike protein-specific IgG were higher than anti-nucleocapsid antibodies. Further, functional assays demonstrated that elevated antibody levels were correlated with increased neutralization of the virus. Using BCR receptor profiling, the authors also found that SARS-CoV-2 RBD-specific clones acquired somatic mutations and incorporated VDJ rearrangements over time. These are part of a germinal center-dependent immune response and often predict the presence of long-lasting memory B cells.
Together, these results provide insight into the B cell response following infection and indicate that a SARS-CoV-2 specific response was still present 6 months following infection. Additional analysis of the B cell receptors indicates that this response may be long-lived.
The development of vaccines and therapeutics for infectious diseases, like SARS-CoV-2, relies on characterizing the immune response, in which B and T cells play a critical role. We provide comprehensive solutions to help you understand the complete immune response to infection. In addition to our TotalSeq™ reagents for single-cell multiomics, our scientists have created recombinant proteins, antibodies, ELISAs, and multiplex assays to accelerate COVID-19 research.
 Login/Register
Login/Register 






Follow Us