TotalSeq™-A Antibodies and Cell Hashing with 10x Single Cell 3' Reagent Kit v3.1 (Dual Index) Protocol
This is the TotalSeq™-A antibodies and cell hashing with 10x Single Cell 3’ Reagent Kit v3.1 (dual Index) protocol. We update our protocols on a regular basis to provide improved guidance on use of our reagents and to enhance the user experience.
Buyer is solely responsible for determining whether Buyer has all intellectual property rights that are necessary for Buyer's intended uses of the BioLegend TotalSeq™ products. For example, for any technology platform Buyer uses with TotalSeq™, it is Buyer's sole responsibility to determine whether it has all necessary third-party intellectual property rights to use that platform and TotalSeq™ with that platform.
The protocol below is intended for customers who are using TotalSeq™-A antibodies and cell hashing reagents with the 10x Single Cell 3’ Reagent Kit v3.1 Dual Index kit.
For customers using TotalSeq™-B or TotalSeq™-C antibodies, please refer to our TotalSeq™-B or C protocol.
Please read the entire protocol below and the 10x Genomics user guide and CG000317 for v3.1 reagents, before starting your experiments.
Commonly used abbreviations and terminology:
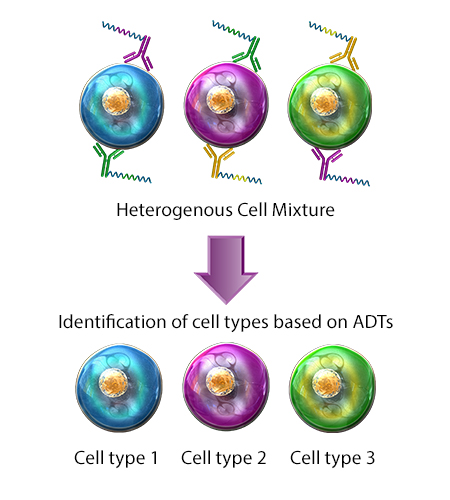
- ADT - Antibody Derived Tag. Refers to a TotalSeq™ DNA-barcoded oligonucleotide that is directly conjugated to a specific antibody clone of interest. Each antibody clone was raised against a specific extracellular protein epitope and can be used to characterize the expression of that surface antigen on cells. Thus, ADTs serve as DNA tags used to catalog and quantify distinct surface protein expression levels.
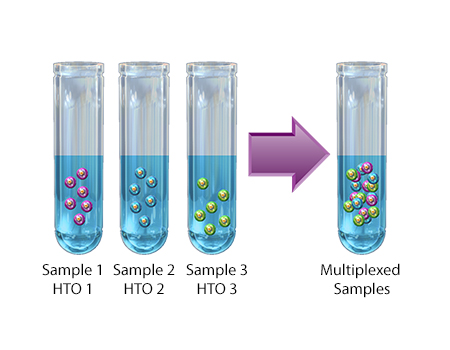
- HTO – Hash Tag Oligonucleotide. Refers to a TotalSeq™ DNA-Barcoded Oligonucleotide that is directly conjugated to a cocktail of two independent antibody clones that are specific for surface proteins known to be ubiquitously expressed on various cell types. The intention of an HTO is to enable researchers to load multiple samples onto a single lane of a 10x chip and maintain the ability to determine sample origin.
- CITE-Seq - Cellular Indexing of the Transcriptome and Epitopes by Sequencing. This application was first described by Stoeckius et al. (Nat Methods 14, 865–868 (2017)) of the NY Genome Center as they used antibodies coupled with oligonucleotides to simultaneously measure proteins and RNA at a single-cell level.
- Cell Hashing – Application where oligo-tagged antibodies against ubiquitously expressed surface proteins uniquely label cells from distinct samples, which can be subsequently pooled. By sequencing these tags alongside the cellular transcriptome, users can assign each cell to its original sample.
Learn more by reading our TotalSeq™/Proteogenomics FAQ.
Multiomic Cytometry
Also known as CITE-seq, multiomic cytometry is a single-cell analysis technique in which cells are stained with antibodies conjugated to short DNA oligos. These oligos contain a sequence barcode, also known as an antibody derived tag (ADT), which is used to identify targeted cellular surface epitopes. Protein and RNA expression can then be characterized in single cells, simultaneously. CITE-Seq can be paired with cell hashing to allow sample multiplexing during sequencing.
Compatibility:
Cell Hashing
TotalSeq™– A
TotalSeq™– B
TotalSeq™– C
Cell Hashing
A technique used to stain cells for multiplexing single-cell partitioned samples during sequencing. Each cell hashing reagent contains a mixture of two distinct monoclonal antibodies targeting distinct ubiquitous cellular surface epitopes. Each monoclonal antibody is conjugated with a short DNA oligo containing the same sequence barcode, also known as hashtag oligo (HTO). HTOs are used to correlate tagged cells with their sample of origin, as they can be mixed together after staining. Cell hashing is compatible with multiomic cytometry and other single-cell analysis techniques.
Compatibility:
Multiomic Cytometry
TotalSeq™– A
TotalSeq™– B
TotalSeq™– C
Reagent and Instrument List
Primers used in sequencing library construction
Must be ordered from 3rd party vendors prior to starting the protocol:
cDNA amplification Step
- ADT additive primer (0.2µM Stock) &/Or
- HTO additive primer v2 (0.2µM Stock)
ADT &/or HTO Library Construction Step
- Dual Index ADT (DI_ADTx) i5/i7 Primer Pairs (10µM Stock) &/Or
- Dual Index HTO (DI_HTOx) i5/i7 Primer Pairs (10µM Stock)
(See notes at the end of the protocol for further details on primer sequences.)
For a full list of sequences, view the primer sequence table below or download this table.
For cell surface staining:
- TotalSeq™-A antibodies and/or TotalSeq™-A hashtag reagents
- Biotinylated antibody and TotalSeq™-A oligo-barcoded streptavidin (optional)
- Human TruStain FcX™ (Fc Receptor Blocking Solution) (BioLegend, Cat# 422301/422302)or TruStain FcX™ PLUS (anti-mouse CD16/32) (BioLegend, Cat# 156603/156604)
- Phosphate Buffered Saline (PBS) (BioLegend, Cat# 926201 or equivalent)
- Cell Staining Buffer (BioLegend, Cat# 420201)
- 12 x 75mm Falcon™ Round-Bottom Polystyrene Tubes (Fisher Scientific, Cat# 14-959-1A or equivalent)
- Flowmi™ Cell Strainer (Bel-Art, H-B Instrument, Cat# H13680-0040)
Optional
- Curiox Laminar Washing
- Some users may be using the Curiox Laminar Wash system, which has equivalent performance to centrifugation-based washing.
- Corning™ ThermalTray™ Thermo-conductive Platforms (Product Number 432074)
For library preparation:
- Quantabio sparQ HiFi PCR Master Mix (2X) (Quantabio, Cat# 95192-250) or KAPA HiFi HotStart ReadyMix (2X) (Kapa Biosystems, Cat# KK2601)
- Quantabio sparQ PureMag Beads (Quantabio, Cat# 951960) or SPRIselect reagent (Beckman Coulter, Cat# B23317)
- 4200 TapeStation (Agilent Technologies, Cat# G2991A)
- DNA High Sensitivity D1000 and High Sensitivity D5000 (Agilent, Cat# 5067-5584/5067-5592)
- Qubit™ 3 (Thermo Fisher Scientific, Cat# Q33226)
- Qubit™ dsDNA HS Assay Kit (Thermo Fisher, Cat# Q32854/Q32851)
- ADT additive primer (0.2µM Stock) and/or HTO additive primer v2 (0.2 µM Stock) (See notes at the end of the protocol for further details on primer sequences.)
- Dual Index ADT i5/i7 Primer Pairs (DI_ADTx) (10µM Stock) and/or Dual Index HTO i5/i7 Primer Pairs (DI_HTOx) (10 µM Stock)
Other essential reagents:
- Nuclease-free Water (Thermo Fisher, Cat#AM9937)
- Ethanol (Sigma, Cat# E7023-500ML)
- Nuclease-Free Pipette Tips (e.g. Thermo Fisher Scientific AM12650, AM12660 or equivalent)
- TempAssure PCR 8-strips (USA Scientific, Cat# 1402-4700)
- PCR Thermocycler (Bio-Rad, T100™ Thermal Cycler)
- Countess™ II FL Automated Cell Counter (ThermoFisher, Cat# AMQAF1000)
Researchers are advised to validate equivalent products when substituting for the above recommendations.
Protocol
I) Cell labeling
- Prepare cell suspensions.
- This protocol has been optimized using fresh human PBMCs isolated using Ficoll gradients and mouse splenocytes prepared using mechanical dissociation. If using cells isolated from whole lysed blood or other sample types, users may need to optimize staining concentrations.
- BioLegend has not tested this protocol using single cell suspensions derived from enzymatically digested tissue. Enzymatic digestion may result in cleavage of epitopes and result in reduced staining with TotalSeq™ antibodies. Optimization of staining conditions and concentrations may be required.
- Assess Cell Viability. Carefully count all cells to ensure accurate quantitation and assess cell viability. Ideal cell viability is ≥ 95%.
- Low cell viability is associated with generation of poor data and is not ideal for single cell experimentation. If low cell viability is observed, users may need to enrich live cells or repeat cell suspension preparation.
- Contact Technical Services with any questions regarding cell viability. BioLegend uses a Countess II for counting and assessing cell viability, however other methods for assessing cell viability are suitable. For more information about the protocol used by BioLegend, see the following link, details can be found under “PBMC viability assessment—general methods”.
- Dilute cells in appropriate volume prior to staining.
- If working with human cells, dilute 1 million cells in 45 μL of Cell Staining Buffer in 12 x 75mm flow cytometry tubes.
- If working with mouse cells, dilute 1 million cells in 49.5 μL of Cell Staining Buffer in 12 x 75mm flow cytometry tubes.
- Block cells.
- Add 5 µL of Human TruStain FcX™ Fc Blocking reagent or 0.5 µL of TruStain FcX™ PLUS (anti-mouse CD16/32) antibody. The final blocking volume should be 50 µL.
- Incubate for 10 min at 4°C.
- While cells are incubating in Fc Block, proceed to step 5.
- Prepare antibody pool using titrated amounts (up to 1 µg) of each TotalSeq™, Cell Hashing, and/or biotinylated antibody. For more information regarding TotalSeq™ antibody concentrations, please reach out to BioLegend Tech Services.
- When performing dual staining with cell hashing antibodies and TotalSeq™ antibodies, we recommend adding cell hashing antibodies into each respective sample’s TotalSeq™ antibody pool. If sequentially staining samples, stain with cell hashing antibodies first and then pool your hashtag labeled samples for TotalSeq™ antibody staining. Please note that in some cases cell recovery may be diminished.
- If using biotinylated antibodies, we recommend staining with your primary antibody first followed by staining with streptavidin TotalSeq™ conjugates. Do not stain with more than 1 unique biotinylated antibody for detection.
- If antibody cocktail volume is less than 50 µL, add Cell Staining Buffer up to 50 µL, then centrifuge the antibody pool at 14,000 x g at 2 – 8°C for 10 minutes before adding to the cells. If volume of pool is above 50 µL, no volume adjustment is necessary.
- If using an antibody cocktail larger than 50 µL, contact Tech Services or your local Technical Applications Scientist for protocol guidance before proceeding with this protocol.
- Carefully pipette out the prepared antibody pool, avoiding the bottom of the tube, and add the TotalSeq™ antibody cocktail to the 50 µL blocked cell suspension.
- Note for Curiox Laminar Wash system users: Transfer 80μL* of cell suspension to one well of the Curiox Wash plate and place on the Thermal Tray** on ice.
i) *80μL is the maximum volume of cells that can be used with Curiox Laminar Wash system.
ii) **Either ThermalTray or traditional incubation methods can be used.
- Note for Curiox Laminar Wash system users: Transfer 80μL* of cell suspension to one well of the Curiox Wash plate and place on the Thermal Tray** on ice.
- Incubate for 30 minutes at 4°C.
- Note for Curiox Laminar Wash system users: Perform the following after step 8, transfer Curiox Wash Plate to the Curiox Laminar Wash System and wash using the following parameters (Flow Rate: 10, # of cycles: 25). Remove Curiox Wash Plate from the system and add 40μL of wash buffer to the well containing the washed cells. Resuspend by pipetting gently.
- Proceed to step 11.
- Add 3 mL of Cell Staining Buffer and spin at 4°C for 5 minutes at 400 x g -600 x g depending on your sample type. Repeat wash 2 more times for a total of 3 washes.
- BioLegend recommends the use of a swing bucket centrifuge as centrifuging with fixed angle rotors may lead to “smearing” of the cell pellet, which may result in cell loss. Contact technical service or local field representative if you have any questions.
- BioLegend recommends manually pouring out the supernatant, being careful not to disrupt the cell pellet. Between 50- 150 μL of residual supernatant will remain in the tube after decanting, which is taken into consideration in step 11 of this protocol. Do not try to forcefully remove any remaining liquid as this will disrupt the cell pellet and result in cell loss.
- If using a biotinylated primary antibody, incubate the stained cells with the appropriate oligo barcoded streptavidin at the recommended amount specified on the product technical datasheet for 20 minutes. Repeat step 9, then proceed to step 11.
- Add 200 μL of Cell Staining Buffer to the cells for an approximate final volume of 250-350 µL.
- Slowly filter cells through 40 µm Flowmi™ Cell Strainer.
- Note: 40 µm Flowmi™ Cell Strainer may be too small for some sample types.
- Verify cell concentration and viability after filtration.
- Note: If using the 10x Genomics Chromium Controller for single cell partitioning, we highly recommend determining cell viability. Ideally the viability should be >90% after filtration for optimal capture rate. The presence of a large number of non-viable cells can decrease the efficiency of cell partitioning and recovery within the 10x Genomics Chromium chip.
- Adjust cell concentration using PBS according to the input requirements of your single cell partitioning platform.
II) Run 10x Genomics single cell 3' v3.1 assay as described through Post Gem-RT Cleanup – Dynabeads (step 2.1). 10x Genomics Documents CG000317 for v3.1.
- At cDNA amplification step (Step 2.2), use the following table to prepare cDNA amplification mix:
ADT 1 rxn (µL) HTO 1 rxn (µL) ADT + HTO 1 rxn (µL) Amp Mix 50 50 50 cDNA Primers* 15 15 15 ADT Additive Primer (0.2 µM stock) 1 0 1 HTO Additive Primer v2 (0.2 uM stock) 0 1 1 Total 66 66 67 *included with 10x Genomics 3’ kit (PN 2000089), different from Feature cDNA primers 2.
- Add amplification mix to 35 µL of the sample.
- Mix by pipetting 15x with the pipette set to 90 µL. Centrifuge briefly.
- Proceed to the 10x User Guide CG000317 Rev B step 2.2d (thermal cycler protocol) of the cDNA Amplification.
Important: Follow steps 2.3A and 2.3B exactly to separate ADTs/HTOs from cDNA. Continue to use 70 µL of sparQ or SPRI beads in step 2.3B.
III) ADT and HTO dual index library preparation
Important: Use the DI_ADTx / DI_HTOx primer pairs specified in the primer table, do not mix primer pairs.
Important: For samples that contain both ADTs and HTOs, you will need to perform two separate reactions when preparing the dual index libraries, one for the ADT library and one for the HTO library. Use 5 µL of the “purified ADT/HTO fraction” from step 2.3B of the 10x Genomics protocol for each reaction and proceed with preparing the reaction mix using the table below.
- At Cell Surface Protein Library Construction (Step 4.1) in the 10x user guide CG000317, use the following table to prepare the Sample Index PCR Mix.
ADT HTO Volume (µL) Purified ADT/HTO fraction Purified ADT/HTO fraction 5 DI_ADTx i5 primer (10 µM stock) DI_HTOx i5 primer (10 µM stock) 2.5 DI_ADTx i7 primer (10 µM stock) DI_HTOx i7 primer (10 µM stock) 2.5 2X QuantaBio or Kapa Hifi Master Mix 2X QuantaBio or Kapa Hifi Master Mix 50 RNAse-free water RNAse-free water 40 Total 100 - Incubate in a thermal cycler with the following protocol:
ADT 98°C 2min 98°C 20sec 14 - 15 Cycles 60°C 30sec 72°C 20sec 72°C 5min 4°C hold HTO 98°C 2min 98°C 20sec 13 - 15 Cycles 64°C 30sec 72°C 20sec 72°C 5min 4°C hold - Perform post ADT/HTO library amplification clean-up.
- Add 120 µL sparQ or SPRIselect Reagent (1.2X) to each sample.
- Incubate 5 min at room temperature.
- Place on the magnet in its High position until the solution clears.
- Carefully remove and discard the supernatant.
- Place tubes on magnet in its High position. Wash the pellet twice with 200 µL 80% ethanol.
- Centrifuge briefly. Place on the magnet Low. Remove remaining ethanol.
- Remove from the magnet. Add 40.5 µL water.
- Incubate 2 min at room temperature.
- Place on the magnet in its Low position until the solution clears.
- Transfer 40 µL to a new tube strip. Store at 4°C for up to 72 h or at −20°C for long-term storage.
- ADT/HTO libraries are now ready to be sequenced. Quantify libraries by standard methods (QuBit, BioAnalyzer, qPCR). ADT and HTO libraries will be around 230 bp (Figure 1).
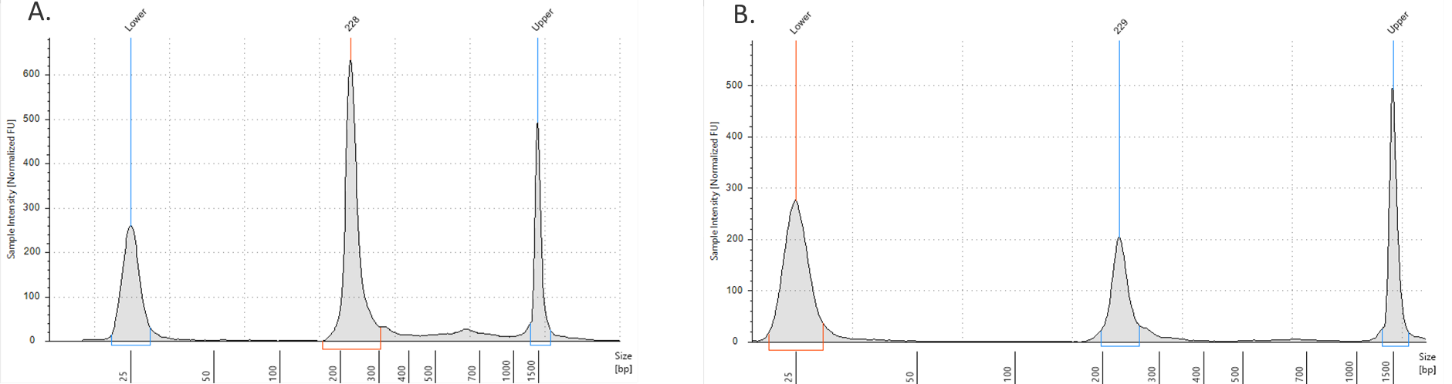
Figure 1. ADT/HTO library verification on the TapeStation D1000 tape. A) ADT Library product (~230 bp). B) HTO library product (~230 bp).
IV) Sequencing CITE-seq libraries:
To obtain sufficient read coverage for both libraries, we typically sequence ADT libraries in 5-10% of a lane and the cDNA library fraction at 90% of a lane (HiSeq2500 Rapid Run Mode Flow Cell). See table below for sequencing depth recommendations.
|
Library Type |
Minimum Sequencing Depth |
|---|---|
|
3' Gene Expression Library |
20,000-50,000 |
|
Cell Surface Protein Library <100 ADT panel |
5,000 |
|
Cell Surface Protein Library ≥100 ADT panel |
10,000 |
|
Cell Hashing Libraries |
500 |
TotalSeq™ A – Dual Index Read Length Requirements
Scenario 1. Sequencing Antibody (ADT &/or HTO) Library and Gene Expression GEX) Library together.
| Read1 | Index1 (i7) | Index2 (i5) | Read2 | |
| Number of cycles | 28 | 10 | 10 | 90 |
Scenario 2. Sequencing Antibody (ADT &/or HTO) Library only
| Read1 | Index1 (i7) | Index2 (i5) | Read2 | |
| Number of cycles | 28 | 10 | 10 | 25 |
Notes regarding oligonucleotide sequences:
TotalSeq™ antibodies:
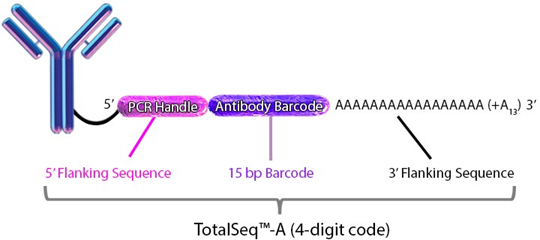
Each clone is barcoded with a unique oligonucleotide sequence. These contain standard small TruSeq RNA read 2 sequences and can be amplified using Dual Index ADT i5/i7 Primer Pairs (DI_ADTx) to obtain Illumina and 10x GEX dual-indexing compatible sequencing libraries): CCTTGGCACCCGAGAATTCCAAACAAGACCCTTGAGBAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA*A*A.
Please visit https://www.biolegend.com/de-at/totalseq for detailed information:
Oligos required for ADT and HTO library amplification:
- PAGE purification is the preferred method when ordering primers.
- The phosphorothioate bonds in the primer renders the oligonucleotide resistant to nuclease degradation.
- A unique Illumina index should be used for each 10x Genomics sample lane used within one sequencing lane.
cDNA Primers
- ADT cDNA PCR additive primer: 5’CCTTGGCACCCGAGAATT*C*C
- HTO cDNA PCR additive primer: v2 5’GTGACTGGAGTTCAGACGTGTGCTCTTCCGAT*C*T
Sequencing Library Primers
- Dual Index ADT i5/i7 Primer Pairs (DI_ADTx) (for ADT amplification; see table 1 for full sequences)
[DI_ADT1 i5] - 5’AATGATACGGCGACCACCGAGATCTACAC-[10 bp index]-ACACTCTTTCCCTACACGACGC*T*C
[DI_ADT1 i7] - 5’CAAGCAGAAGACGGCATACGAGAT-[10 bp index]-GTGACTGGAGTTCCTTGGCACCCGAGAATTC*C*A
- Dual Index HTO i5/i7 primer pairs (DI_HTOx) (for HTO amplification; see table 1 for full sequences)
(DI_HTO1) i5 - 5’AATGATACGGCGACCACCGAGATCTACAC-[10 bp index]-ACACTCTTTCCCTACACGACGC*T*C
(DI_HTO1) i7 - 5’CAAGCAGAAGACGGCATACGAGAT-[10 bp index]GTGACTGGAGTTCAGACGTGTGCTCTTCCGAT*C*T
* indicates a phosphorothioate bond
B indicates C or G or T; not A nucleotide
Primers Used for Sequencing Library Construction:
| Index | Full Sequence 5’ -> 3’ | SampleSheet Index1 |
SampleSheet Index2 (forward strand workflow) |
SampleSheet Index2 (Reverse complement workflow) |
|
| DI_ADT1 | i5 | AATGATACGGCGACCACCGAGATCTACACCGTAGAGCAGACACTCTTTCCCTACACGACGC*T*C | TCTACTCGCG | CGTAGAGCAG | CTGCTCTACG |
| i7 | CAAGCAGAAGACGGCATACGAGATCGCGAGTAGAGTGACTGGAGTTCCTTGGCACCCGAGAATTC*C*A | ||||
| DI_ADT2 | i5 | AATGATACGGCGACCACCGAGATCTACACGACGAGATGCACACTCTTTCCCTACACGACGC*T*C | CTCTGCGCTA | GACGAGATGC | GCATCTCGTC |
| i7 | CAAGCAGAAGACGGCATACGAGATTAGCGCAGAGGTGACTGGAGTTCCTTGGCACCCGAGAATTC*C*A | ||||
| DI_ADT3 | i5 | AATGATACGGCGACCACCGAGATCTACACGATGGCGAACACACTCTTTCCCTACACGACGC*T*C | TCGCAGCTTC | GATGGCGAAC | GTTCGCCATC |
| i7 | CAAGCAGAAGACGGCATACGAGATGAAGCTGCGAGTGACTGGAGTTCCTTGGCACCCGAGAATTC*C*A | ||||
| DI_ADT4 | i5 | AATGATACGGCGACCACCGAGATCTACACCGAACGATGGACACTCTTTCCCTACACGACGC*T*C | CTAGTCGCCT | CGAACGATGG | CCATCGTTCG |
| i7 | CAAGCAGAAGACGGCATACGAGATAGGCGACTAGGTGACTGGAGTTCCTTGGCACCCGAGAATTC*C*A | ||||
| DI_HTO1 | I5 | AATGATACGGCGACCACCGAGATCTACACCGAAGGTCAGACACTCTTTCCCTACACGACGC*T*C | GCTTCACGCT | CGAAGGTCAG | CTGACCTTCG |
| I7 | CAAGCAGAAGACGGCATACGAGATAGCGTGAAGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGAT*C*T | ||||
| DI_HTO2 | I5 | AATGATACGGCGACCACCGAGATCTACACGAGTAACGGCACACTCTTTCCCTACACGACGC*T*C | CTCAGGTCTC | GAGTAACGGC | GCCGTTACTC |
| I7 | CAAGCAGAAGACGGCATACGAGATGAGACCTGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGAT*C*T | ||||
| DI_HTO3 | I5 | AATGATACGGCGACCACCGAGATCTACACTGCAAGGACGACACTCTTTCCCTACACGACGC*T*C | TCTCTGCGAC | TGCAAGGACG | CGTCCTTGCA |
| I7 | CAAGCAGAAGACGGCATACGAGATGTCGCAGAGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGAT*C*T | ||||
| DI_HTO4 | I5 | AATGATACGGCGACCACCGAGATCTACACAATGGACGGCACACTCTTTCCCTACACGACGC*T*C | CTCGACGCTT | AATGGACGGC | GCCGTCCATT |
| I7 | CAAGCAGAAGACGGCATACGAGATAAGCGTCGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGAT*C*T |
* Note: for more Dual Index ADT i5/i7 Primer Pairs (DI_ADTx) and Dual Index HTO (DI_HTOx) i5/i7 Primer Pairs, please refer to this table.
Representative Data and Troubleshooting
My ADT/HTO library contains a large peak at ~140-180bp
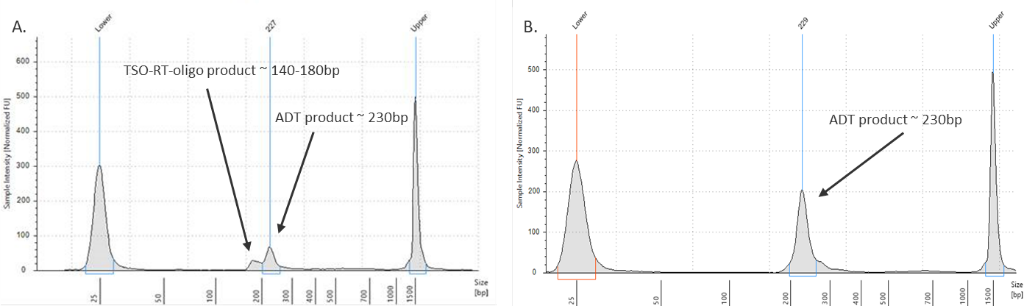
Figure 2. ADT library verification.
(Left graph) A TSO-RT-oligo product (~140 - 160 bp) can be amplified during the ADT PCR by carryover primers from cDNA amplification. The product will not cluster but will interfere with quantification. Sequential 2X sparQ or SPRI purification of the ADT fraction after cDNA amplification reduces carryover of primers from cDNA amplification, and minimizes the amplification of this product during ADT library amplification. To further enrich for ADT specific product the purified ADT library can be reamplified for 3 additional cycles with ADT specific primer sets or P5/P7 generic primers. (Right graph) A clean ADT library will contain a predominant single peak at around 230 bp.
My ADT/HTO library contains a large peak at ~400bp
Overamplification of a library can lead to depletion of available primers and/or dNTPs resulting in self-priming of PCR products by their P5 and/or P7 adapters. This can lead to the production of “daisy-chains” or “bubble products”. These products consist of essentially 2 ADT or HTO barcode sequences attached to one another in 1 long oligo tag that is twice as long as the original oligo tag; these products appear as peaks at approximately 400 - 440 bp. These peaks can be more or less pronounced.
The larger peak is perfectly acceptable to sequence. However, it is difficult to quantify these libraries to titrate for sequencing. This error can be corrected by performing another PCR reaction using generic P5/P7 primers (not used in the protocol) for one or two cycles on the 1.2x sparQ or SPRI cleaned up product. The PCR reaction will convert the larger product to the desired size product partially or completely as the major peak.
 Login / Register
Login / Register 







Follow Us