When working with precious samples that may have less than ideal sample viability or quality, meaningful and rich single-cell multiomics data is still achievable. Explore our poster or read below to discover how BioLegend and Q2 Solutions collaborated to demonstrate this on clinically relevant Human cryopreserved samples.
Cellular Indexing of Epitopes and Transcriptomes by Sequencing (CITE-seq) is a recent technique that has seen wide adoption in the research communities. However, technical guidance as well as vendor support on the use of the technique along with associated reagents and platforms has been limited when experimental conditions are not ideal regarding sample quality (age, viability, cell recovery, etc). Non-ideal sample conditions are best avoided where practical, however, the reality of clinical research as well as the limitation of accessibility to disease or population-relevant sample types may limit the control an investigator has over some of these sample attributes in practice.
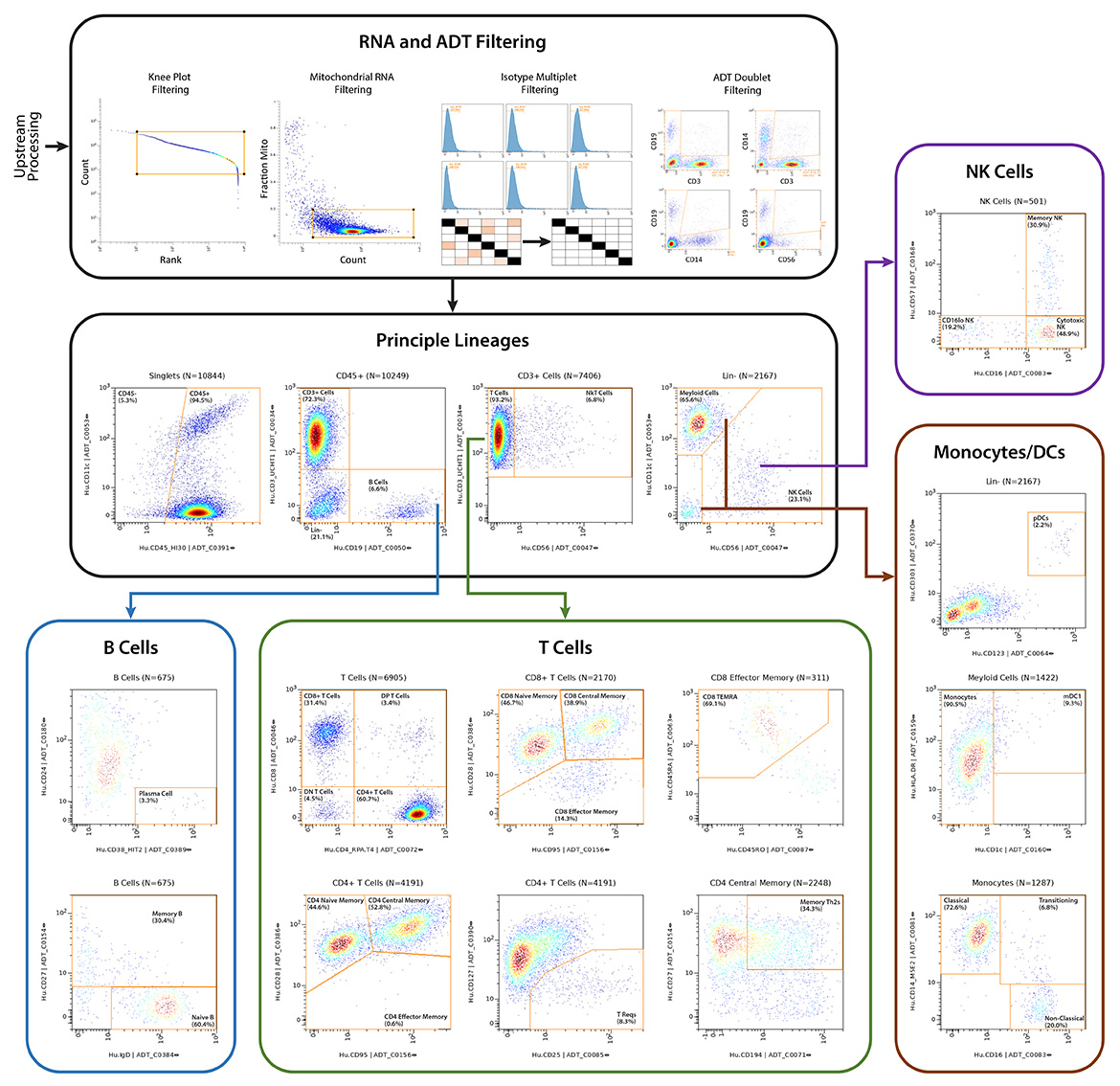
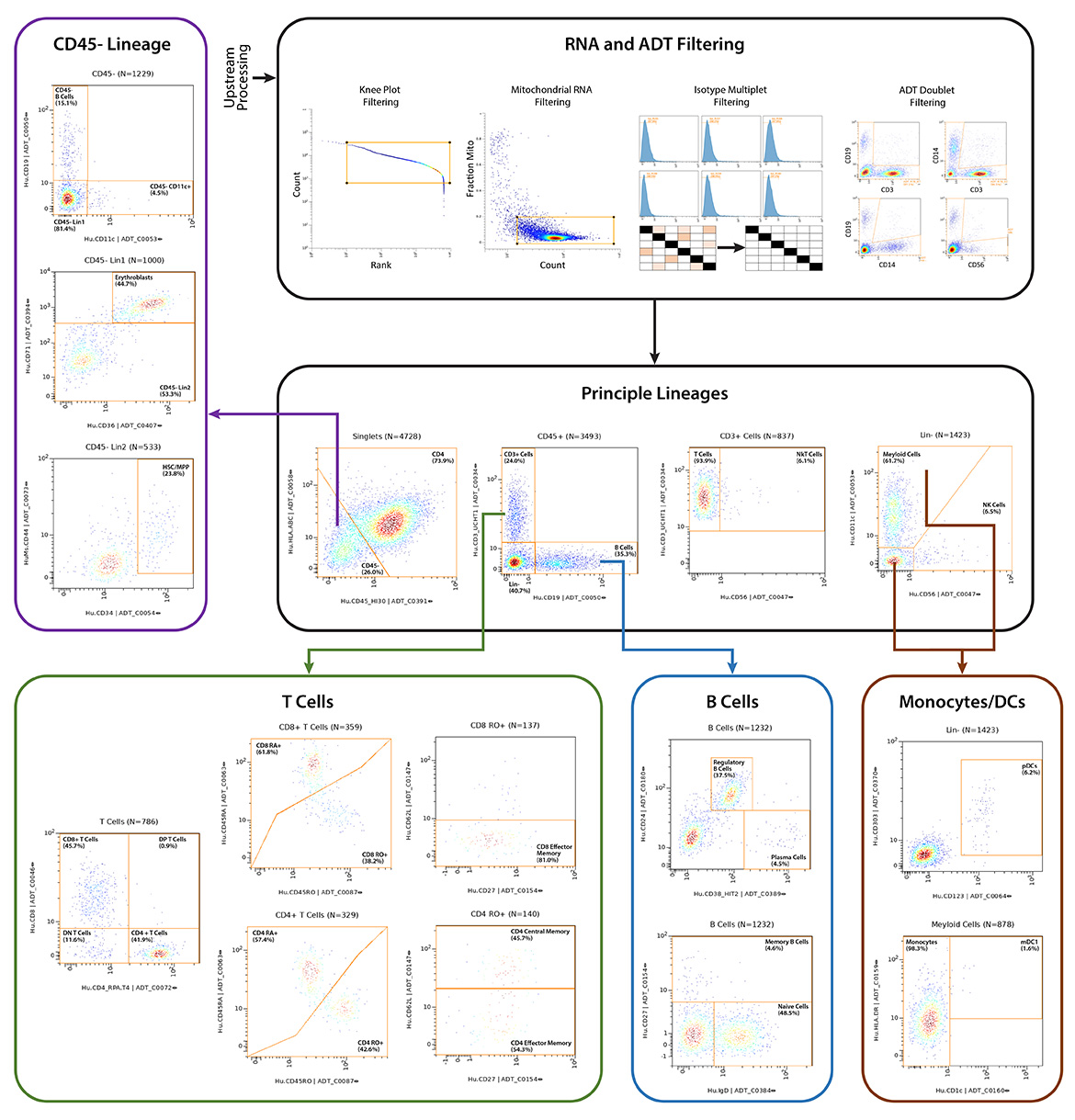
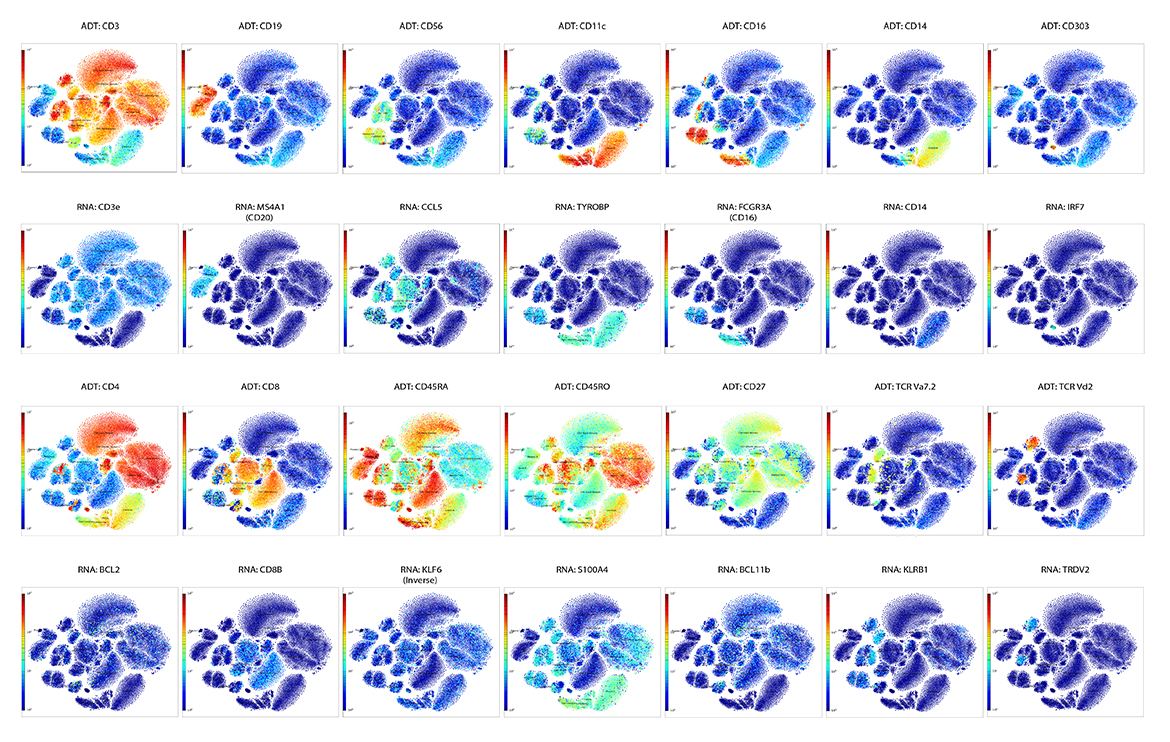
Q2 Solutions and BioLegend engaged in a strategic collaboration to demonstrate the rigor and reproducibility of Q2 Solutions research service offerings on the 10x Genomics product ecosystem using two common human sample types of high clinical relevance for human disease: cryopreserved Peripheral Blood Mononuclear Cells (cPBMCs) and cryopreserved Bone Marrow Mononuclear Cells (cBMMCs). Several matched samples from three donors for both cPBMCs and cBMMCs were acquired and run in duplicate at both BioLegend and Q2 Solutions. All samples were stained using BioLegend TotalSeq™-C Universal Antibody Cocktails (PN 399905) and additional TotalSeq™-C antibodies against human CD235ab, CD90, CD34, CD10, CD135, CD117, CD206, CD271, CD70, CD9, CD200, and, CD74.
cBMMCs can be technically challenging to work with, especially if specimen harvesting and processing is originating in a community setting, and, archival samples are several years old. cBMMCs were processed using identical procedures across both physical locations to investigate intra-site reproducibility.
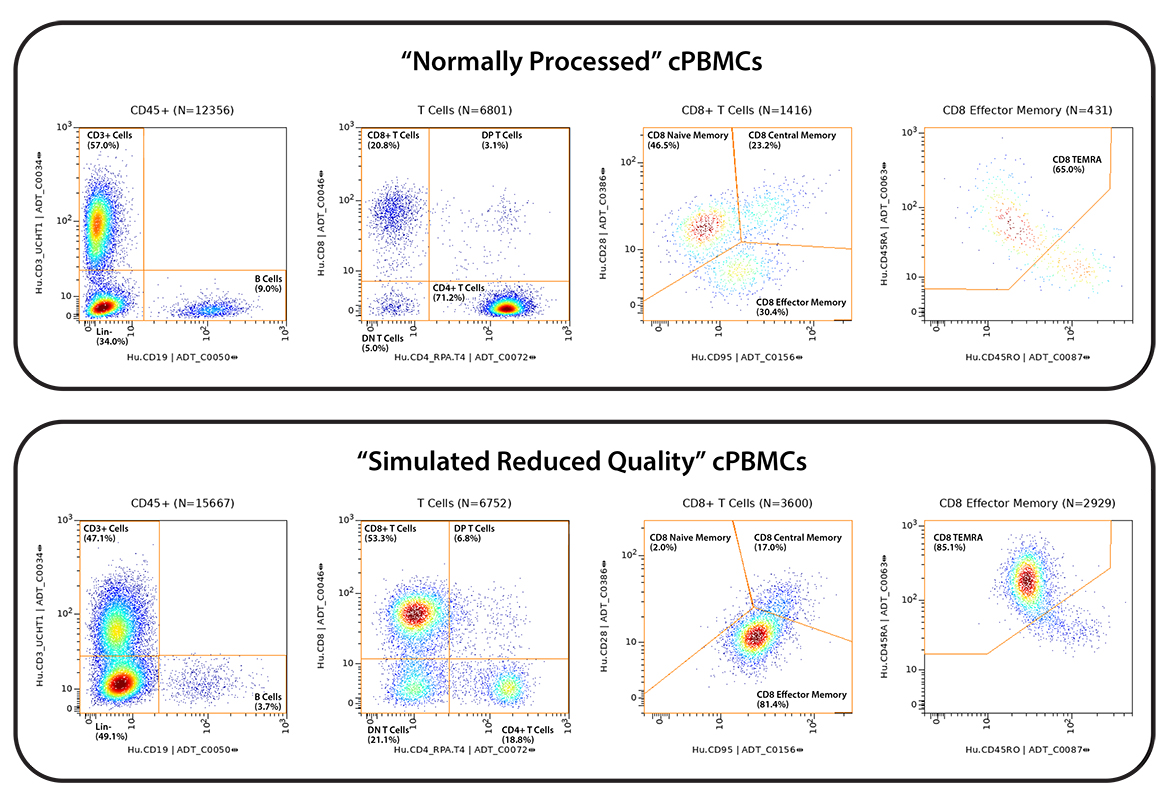
cPBMCs on the other hand are routinely collected across clinical activities in the research setting. Very high-quality cPBMCs can be obtained, however, quality can be highly variable depending on source, specimen, and, handling (storage, time, operator, etc). Thus, simulated low-quality and high-quality cPBMCs were created by temperature shocking ½ of the matched samples between study sites.
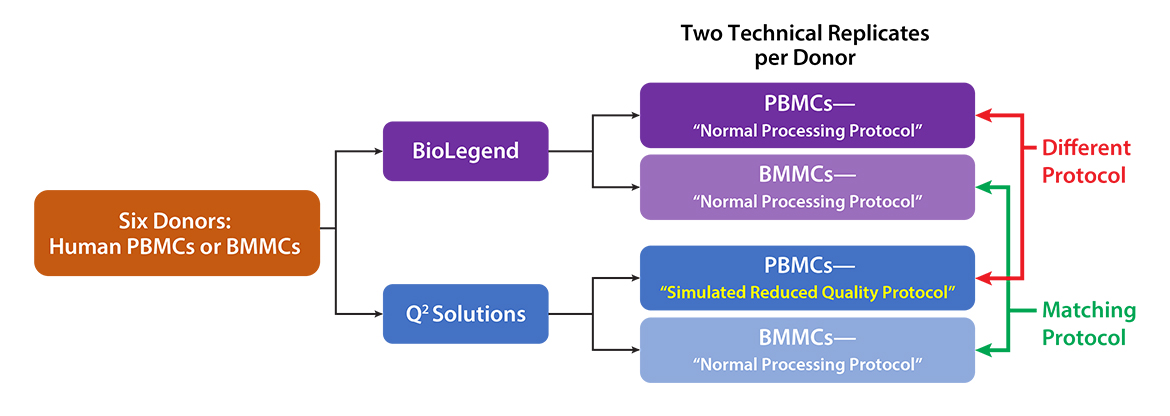
Figure 1 – Study design. All six donors were run at both study sites, in duplicate. All practices were matched except for the introduction of a “cold shock” in the PBMCs at the Q2 study site to simulate reduced sample quality.
 Login/Register
Login/Register 







Follow Us